* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download SALSA MLPA KIT ME003-A1 Tumor suppressor-3 - MRC
Human genome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
DNA supercoil wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetic clock wikipedia , lookup
Genomic library wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Microevolution wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Oncogenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
DNA methylation wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
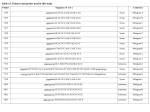
MRC-Holland Description version 04; 05-02-2010 ® MLPA SALSA MLPA KIT ME003-A1 Tumor suppressor-3 Lot 0608 This SALSA® MLPA® kit is for basic research! This kit enables you to detect aberrant methylation of CpG islands upstream of genes for which an altered methylation status in one or more types of tumours has been reported in literature. In case interesting results are obtained by users, it is possible to develop methylation kits specific for a certain tumour in collaboration with MRC-Holland. Interpretation of results obtained with this product can be complicated. MRC-Holland cannot provide assistance with data interpretation. Aberrant methylation of CpG-islands has been shown to be associated with transcriptional inactivation of tumour suppressor genes in a wide spectrum of human cancers. CpG-islands are located in or near the promoter region or other regulatory regions of approximately 50% of human genes. This ME003-A1 MS-MLPA probemix contains 27 MS-MLPA probes which detect the methylation status of promotor regions of 18 different tumour suppressor genes. These tumour suppressor genes are frequently silenced by methylation in tumours, but are unmethylated in blood-derived DNA of healthy individuals. In addition, 13 reference probes are included which are not affected by HhaI digestion. Besides detecting aberrant methylation, all 40 probes present will give information on copy number changes in the analyzed sample. The MLPA reaction requires as little as 20 ng of human DNA and can be used on a variety of DNA samples, including those derived from paraffin-embedded tissues. Please note that each MS-MLPA reaction generates two samples that need analysis by capillary electrophoresis: one undigested sample for copy number detection and one digested sample for methylation detection. More information about MS-MLPA can be found on page 2 and in the MS-MLPA protocol. The MS-MLPA probes in this ME003-A1 probemix detect sequences in promoter regions of tumor suppressor genes that are unmethylated in most blood-derived DNA samples. Upon digestion, the peak signal obtained in unmethylated samples will be very small or absent. In contrast, when tested on in vitro methylated human DNA, these probes do generate a signal. We have no data showing that methylation detected by a particular probe indeed influences the corresponding mRNA levels. This SALSA® MS-MLPA® kit can be used to detect aberrant methylation of one or more sequences of the Tumour suppressor genes. Methylation levels can be different for different tissues. Please use DNA derived from the same type of tissue and purified by the same method as the reference samples. This SALSA® MLPA® kit can be used to detect deletions/duplications of one or more sequences in the above mentioned chromosomal regions in a DNA sample. Heterozygous deletions of recognition sequences should give a 3550% reduced relative peak area of the amplification product of that probe. Note that a mutation or polymorphism in the sequence detected by a probe can also cause a reduction in relative peak area, even when not located exactly on the ligation site! In addition, some probe signals are more sensitive to sample purity and small changes in experimental conditions. Therefore, deletions and duplications detected by MLPA should always be confirmed by other methods. Not all deletions and duplications detected by MLPA will be pathogenic; users should always verify the latest scientific literature when interpreting their findings. Finally, note that most defects in this gene are expected to be small (point) mutations which will not be detected by this SALSA MLPA test. We have no information on what percentage of defects in these genes is caused by deletions/duplications of complete exons. SALSA® MS-MLPA® kits are sold by MRC-Holland for research purposes and to demonstrate the possibilities of the MLPA technique. This kit is not CE/FDA certified for use in diagnostic procedures. SALSA MLPA kits are supplied with all necessary buffers and enzymes. Purchase of the SALSA MLPA test kits includes a limited license to use these products for research purposes. The use of this SALSA® MLPA® kit requires a thermocycler with heated lid and sequence type electrophoresis equipment. Different fluorescent PCR primers are available. The MLPA technique has been first described in Nucleic Acid Research 30, e57 (2002). The MS-MLPA method for the detection of both copy numbers and methylation changes was described in Nucleic Acid Research 33, e128 by Nygren et al. 2005. More information Website : www.mlpa.com E-mail : [email protected] (information & technical questions); [email protected] (for orders) Mail : MRC-Holland bv; Willem Schoutenstraat 6, 1057 DN Amsterdam, the Netherlands SALSA MLPA kit ME003 Tumor suppressor-3 Page 1 of 7 MRC-Holland Description version 04; 05-02-2010 ® MLPA Related SALSA MLPA kits ME001 Tumor suppressor-1: Contains probes for Tumor suppressor genes. ME002 Tumor suppressor-2: Can be used to confirm the results of ME001. ME004 Tumor suppressor-4. Contains probe for Tumor suppressor genes. Methylation-specific MLPA Please note that each MS-MLPA reaction generates two samples that need analysis by capillary electrophoresis: one undigested sample for copy number detection and one digested sample for methylation detection. A modification of the MLPA technique, MS-MLPA allows the detection of both copy number changes and unusual methylation levels of 10-50 different sequences in one simple reaction. MLPA probes for methylation quantification are similar to normal MLPA probes, except that the sequence detected by the MS-MLPA probe contains the sequence recognized by the methylation-sensitive restriction enzyme HhaI. Similar to ordinary MLPA reactions, the MS-MLPA protocol starts with sample DNA denaturation and overnight hybridization. The reaction then is split into two tubes. One tube is processed as a standard MLPA reaction. This reaction provides information on copy number changes. The other tube of the MLPA hybridization reaction is incubated with the methylation-sensitive HhaI endonuclease while simultaneously, the hybridized probes are ligated. Hybrids of (unmethylated) probe oligonucleotides and unmethylated sample DNA are digested by the HhaI enzyme. Digested probes will not be exponentially amplified by PCR and hence will not generate a signal when analyzed by capillary electrophoresis. In contrast, if the sample DNA is methylated, the hemimethylated probe-sample DNA hybrids are prevented from being digested by HhaI and the ligated probes will generate a signal. More information about MS-MLPA can be found in the MS-MLPA protocol. Please note that this product can not be used with an alternative protocol in which the genomic DNA is first digested with Hha1, followed by MLPA reactions on both digested and undigested genomic DNA. Data analysis The ME003-A1 tumour suppressor probemix contains 40 MLPA probes with amplification products between 136 and 481 nt. In addition, it contains 9 control fragments generating an amplification product smaller than 120 nt: four DNA Quantity fragments (Q-fragments) at 64-70-76-82 nt, three DNA denaturation control fragments (D-fragments) at 88-92-96 nt, one X-fragment at 100 nt and one Y-fragment at 105 nt. More information on how to interpret observations on these control fragments can be found in the MLPA protocol. The analysis of MS-MLPA kits consists of two parts: 1) determining copy numbers by comparing different undigested samples (all MLPA kits), and 2) determining methylation patterns by comparing each undigested sample to its digested counterpart (MS-MLPA kits only). The second part is unique for MS-MLPA kits and serves to semi-quantify the percentage of methylation within a given sample. Intra-sample data normalisation (all samples) For analysis of MLPA results, not the absolute fluorescence values but “intra-normalized” data are used (relative peak areas). The data generated in the digested and undigested sample should first be normalized intra-sample by dividing the signal of each probe by the signal of every reference probe in that sample, thus creating as many ratios per probe as there are reference probes. Subsequently, the median of all these produced ratios per probe should be taken; this is the probe’s Normalisation Constant. This Normalization Constant can then be used for sample to reference sample comparison, both for copy number and digestion determination. Copy numbers determination (comparison between undigested samples) The final probe ratio, or ploidy status, of each probe is calculated by comparing a) the intra-normalized ratio of each probe obtained on the undigested patient sample with b) the average intra-normalized ratio of each probe obtained on the undigested reference samples. Methylation analysis (comparing digested and undigested samples) Methylation status of MS-MLPA probes* is calculated by dividing a) the Normalization Constant of each MSMLPA probe obtained on the digested patient sample by b) the Normalization Constant of each MS-MLPA probe obtained on the corresponding undigested sample. Multiplying this value by 100 gives an estimation of SALSA MLPA kit ME003 Tumor suppressor-3 Page 2 of 7 MRC-Holland Description version 04; 05-02-2010 ® MLPA the percentage methylation. Aberrant methylation can then be identified by comparing the methylation status of one or more MS-MLPA probes in the sample in question to that obtained on reference samples. *Note: An MS-MLPA probe targets a single specific HhaI site in a CpG island; if methylation is absent for a particular CpG-site, this does not necessarily mean that the whole CpG island is unmethylated! For samples containing both tumour and normal cells, MLPA experiments will indicate the average copy number of genes. Data used for normalisation should have been obtained within a single experiment. Only samples from the same tissue and purified by the same method should be compared. Confirmation of most exons deletions and amplifications can be done by e.g. Southern blots, long range PCR, qPCR, FISH. Note that Coffalyser, the MLPA analysis tool developed at MRC-Holland, can be downloaded free of charge from our website www.mlpa.com. This probemix was developed by A. Errami at MRC-Holland. In case the results obtained with this probemix lead to a scientific publication, it would be very much appreciated if the probemix designer could be made a coauthor. Info/remarks/suggestions for improvement: [email protected]. SALSA MLPA kit ME003 Tumor suppressor-3 Page 3 of 7 MRC-Holland ® MLPA Description version 04; 05-02-2010 Table 1. SALSA MS-MLPA ME003-A1 Tumour suppressor-3 probemix Length (nt) 64-70-76-82 88-92-96 100 105 136 142 148 157 + 166 178 184 193 202 211 220 229 238 247 256 265 274 283 292 301 310 319 330 337 346 ٨ 355 364 373 382 391 400 409 418 427 436 445 454 463 472 481 SALSA MLPA probe Hha1 site Chromosomal position Q-fragments: DNA quantity; only visible with less than 100 ng sample DNA D-fragments: Low signal of 88 or 96 nt fragment indicates incomplete denaturation X-fragment: Specific for the X chromosome Y-fragment: Specific for the Y chromosome Reference (DNAH5) probe 08035-L07816 CCND2 probe 03312-L09381 SCGB3A1 probe 03305-L09382 BNIP3 probe 07138-L12958 DLC1 probe 02753-L02202 Reference (PTK2) probe 08236-L08114 DLC1 probe 02754-L02203 HLTF probe 02758-L02207 Reference (MLH3) probe 01245-L00793 SFRP5 probe 09149-L03207 CCND2 probe 03313-L02668 Reference (PAH) probe 02334-L01820 H2AFX probe 08511-L08607 CACNA1G probe 10123-L10466 Reference (BCL2) probe 00587-L00382 SFRP4 probe 03744-L03204 TWIST1 probe 02080-L02886 BCL2 probe 10352-L10890 Reference (NF1) probe 02524-L01955 CACNA1A probe 09055-L09224 Reference (CDK6) probe 03184-L02523 TIMP3 probe 10354-L10892 SFRP5 probe 09148-L12957 Reference (CDH1) probe 02416-L01862 ID4 probe 04497-L03909 H2AFX probe 08509-L08605 Reference (AI651963) probe 01234-L00781 RUNX3 probe 11131-L03905 SFRP4 probe 09147-L03205 Reference (KLK3) probe 00713-L00108 TIMP3 probe 10357-L10895 SCGB3A1 probe 11132-L12956 Reference (ATM) probe 02670-L02137 PRDM2 probe 09146-L02862 TGIF probe 02850-L13256 Reference (CD27) probe 00678-L00124 HLTF probe 09152-L09384 ID4 probe 04496-L03908 Reference (SLC6A5) probe 08946-L09041 RARB probe 10362-L10900 + + + + + + + + + + + + + + + + + + + + + + + + + + + 5p15.2 12p13.3 5q35 10q26.3 8p22 8q24 8p22 3q25.1 14q24.3 10q24.1 12p13.3 12q23 11q23.3 17q21.33 18q21.33 7p14.1 7p21.2 18q21.3 17q11.2 19p13.2 7q21.3 22q12.13 10q24.1 16q22.1 6p22.3 11q23.3 10p14 1p36.11 7p14.1 19q13.33 22q12.13 5q35 11q23 1p36.21 18p11.31 12p13.31 3q25.1 6p22.3 11p15.1 3p24.2 + Results obtained with the BNIP3 probe should be disregarded as this probe probably also recognizes a sequence on chromosome 14. Will be replaced in the next lot. ٨ This probe is not completely digested in DNA samples derived from blood. Note: Please notify us of any mistakes. The identity of the genes detected by the reference probes is available on request: [email protected]. SALSA MLPA kit ME003 Tumor suppressor-3 Page 4 of 7 MRC-Holland ® MLPA Description version 04; 05-02-2010 Table 2. ME003 probes arranged according to chromosomal location Length (nt) SALSA MLPA probe Gene HhaI site Chr & kb to p-telo 373 11131-L03905 RUNX3 + 01-025.1 427 09146-L02862 PRDM2 + 01-013.8 481 » 10362-L10900 RARB + 03-025.4 454 09152-L09384 HLTF + 03-150.2 193 02758-L02207 HLTF + 03-150.2 136 08035-L07816 DNAH5 148 03305-L09382 SCGB3A1 + 05-179.9 409 11132-L12956 SCGB3A1 + 05-179.9 346 04497-L03909 ID4 + 06-019.9 463 04496-L03908 ID4 + 06-019.9 265 03744-L03204 SFRP4 + 07-037.9 382 09147-L03205 SFRP4 + 07-037.9 274 02080-L02886 TWIST1 + 07-019.1 310 03184-L02523 CDK6 184 02754-L02203 DLC1 + 08-013.03 166 02753-L02202 DLC1 + 08-013.03 178 08236-L08114 PTK2 08-141.8 364 01234-L00781 AI651963 10-011.01 211 09149-L03207 SFRP5 + 10-099.5 330 09148-L12957 SFRP5 + 10-099.5 157 + 07138-L12958 BNIP3 + 10-133.6 472 08946-L09041 SLC6A5 11-020.6 418 02670-L02137 ATM 11-107.6 238 08511-L08607 H2AFX + 11-118.4 355 08509-L08605 H2AFX + 11-118.4 220 03313-L02668 CCND2 + 12-004.2 142 03312-L09381 CCND2 + 12-004.2 445 00678-L00124 CD27 12-006.4 229 02334-L01820 PAH 12-101.7 202 01245-L00793 MLH3 14-074.5 337 02416-L01862 CDH1 16-067.4 292 02524-L01955 NF1 17-026.6 247 10123-L10466 CACNA1G 05-013.9 07-092.08 + SALSA MLPA kit ME003 Tumor suppressor-3 17-045.9 (partial) sequence with HhaI site GTGCTTGGGTCTACGGGAATACGCATAACAGCGGCCGTCAGGGCGCCGGGCAGGCGGA GCTGGGTGGTGGCCATTGGGCGACGGCGCAGGGTCAAGGGCCGGGCTCTGGGATCGTG CATGTGCTTTTTCTGGAGTGGAAAAATACATAAGTTATAAGGAATTTAACAGACAGAAAGGCGCACAGAGGAATTT CGGCTCGAAAACGATCCAGGGGAGCCGAGGCGCTCCTCTTGTCATCCCACTCAGCGCCAT GACTGGACTCGCGGCGACTTACCTTTCAGTCGTGCGCTCCTGATCCGGCGCTCGGAATTTG TTGTGGCTTCCTGTTTGGACCTGAACAAAACCGAAGTGGAGGATGCCATTCTTGAAGGGAATCA GGGCCTCGAGGACTTCCTCTTGGCAGGCGCTGGGGCCCTCTGAGAGCAGGCAGGCCCGGCCTT CCAGGAGGGCGGCGAGCTTCATGGCGCGGGGGCTCGGGGCGCGCGGGGAACCT GGGAAATATAGAACAGAATGAACGTGCCTTCTTGAACAGCGCGTCTTTCTTAAGGCACTGGAATCCC GTAGTGGAGGAGGCGCGGTTGTGAGTAGTACCGGGAGTGGGGTGATCCCGGGCTAGGGGAGCGC GAAAGATGAGGGTGGCAGGAAGAGAAGGGCGCTTTCTGTCTGCCGGGGTCGCAGCGCGAGAGGG CGGGATAAATAGGGTCCCGCAATGGCCGTGGCTGGCTGCGCTCCGAGCTGCGGAGTCC CGCAGACTTCTTGCCGCGCTTGCCCTGGGCCGGGCTGCCCGGCTCGTCGCCG GCGTGATTGGACTCCCAGGAGAAGAAGACTGGCCTAGAGATGTTGCCCTTCCCAGGCAGGCTTTTCA GCCAATAGTTGGCGACTTTTCGGCGCTTCCCGCTGGGAACGTGGAGGCCCGTGGGGGAAAC CAAGGATGCGTTGAGGACCGCGAGGGCGCGCGTCTCGGGTGCCGCCGTGGGT TCTGACAGTGACGGCACCATCCCTAACCATTGCGGAGAATATGGCTGACCTAATAGATGGGTACTGCCGG CAATTGCCATTTTTTCCTGACATTCACTGTGGAAATTTGGTGCACGACACTGTTAGGGGAGATCTGT GTCCCGCCCAGCGCCAGGTCTTGCGCCCCTCTCCCGGCTCGGCTCCCCTGCCGCA CGCTCGGCCTGCCAGCCATAGTAGTCGTACTCCTCGCAGCGCGCCGGCGCCCAGTGCAGCGC GGCGCGGACTCCGATCGCCGCAGTTGCCCTCTGGCGCCATGTCGCAGAACGGAGC CCATGACCTATGGCTCTTACCGCTATCCTAACTGGTCCATGGTGCTCGGATGGCTAATGCTCGCCTGTTC GCATATCATAGACCTTGGTAGCAGTCTCTCTTTGCTGTGCCATCCCACTGTCTCTGGTACCCTCATAGGC CCAACCGGAGGCGGGTATTGGAGAAAAGAGCCAATCAGGAGGGCGCAGAGGTGTGTCCTGGGGGCTT GATTGTAGTGTTTAAATCTCGCGCGCTTTACAAGGATTGGCTACATCAGCGGATAGTTGGCAGTCTG CAGGGAGAAAGCCTGGCGAGTGAGGCGCGAAACCGGAGGGGTCGGCGAGGATGCGGGCGAAGGA CTGCATCGGTGTGGCCACGCTCAGCGCAGACACCTCGGGCGGCTTGTCAGCAGATGCAGG GAAAGTCCTGTGGAGCCTGCAGAGCCTTGTCGTTACAGCTGCCCCAGGGAGG CAGTGCCCTGGTTCCCAAGAACCATTCAAGAGCTGGACAGATTTGCCAATCAGATTCTCAG GCGACCTTGTTCTTCCTTTCCTTCCGAGAGCTCGAGCAGAGAGGACTGTGATGAGACAGGATAACAG CTATGAAGGAAGCGGTTCCGAAGCTGCTAGTCTGAGCTCCCTGAACTCCTCAGAGTCAGACAAAGACCAGGAC GGCAATCCGGAATCCTCTGGAGTGGCACTGCAAGCAAATGGATCATTTTGTTGGACTCAATTTCAACT CGGGCGATCCGGAGAGGGGCA- Page 5 of 7 MRC-Holland ® MLPA Description version 04; 05-02-2010 Length (nt) SALSA MLPA probe Gene HhaI site Chr & kb to p-telo 436 02850-L13256 TGIF + 18-003.4 283 10352-L10890 BCL2 + 18-059.1 256 00587-L00382 BCL2 301 09055-L09224 CACNA1A 391 00713-L00108 KLK3 400 10357-L10895 TIMP3 + 22-031.5 319 10354-L10892 TIMP3 + 22-031.5 (partial) sequence with HhaI site AGCGGCGCCCCTCAGAGGAGGTGTCCTCACGCAA CGTTTCTGTGGGAGGCCCTGAAACGCGCGGAGCTTCCCTCTGCCTCCAGGCTTTCCCAGCGAG GTGAAGCGGTCCCGTGGATAGAGATTCATGCCTGTGCCCGCGCGTGTGTGCGCGCGTG TTCTCCTGGCTGTCTCTGAAGACTCTGCTCAGTTTGGCCCTGGTGGGAGCTTGCATC CGGGGCGCAAAGGATGTACAAGCAGTCAATGGCGCAGAGAGCGCGGACCATG TGTGTCACCATGTGGGTCCCGGTTGTCTTCCTCACCCTGTCCGTGACGTGGA CCAGCGCTATATCACTCGGCCGCCCAGGCAGCGGCGCAGAGCGGGCAGCAGGCAG CATCGTGCTCCTGGGCAGCTGGAGCCTGGGGGACTGGGGCGCCGAGGCGTGCACA 18-058.9 + 19-013.4 19-056.05 + Results obtained with the BNIP3 probe should be disregarded as this probe probably also recognizes a sequence on chromosome 14. Will be replaced in the next lot. » A second probe for this gene, recognizing a different CpG-site, is present in the SALSA MLPA kit ME002. The Hha1 sites are marked with grey. Ligation sites are marked with – Note: Complete probe sequences are available on request: [email protected]. Please notify us of any mistakes: [email protected]. SALSA MLPA ME003-A1 Tumour suppressor-3 sample pictures 70000 301,23 60000 164,63 328,60 245,96 155,50 381,32 177,64 50000 193,40 237,51 85,53 40000 105,56 95,96 134,76 182,82 280,00 228,55 91,14 30000 398,80 218,25 209,31 148,66 140,78 354,76 291,35 201,13 452,74 256,39 273,43 264,15 20000 100,71 318,55 345,46 336,97 371,26 483,17 474,85 409,32 426,71 308,82 389,77 418,48 436,48 364,13 443,52 463,55 Dye Signal 10000 0 100 150 200 250 300 Size (nt) 350 400 450 Figure 1. Capillary electrophoresis pattern from a sample of approximately 50 ng undigested human male control DNA analyzed with SALSA MLPA kit ME003-A1 Tumor suppressor-3 (lot 0608). SALSA MLPA kit ME003 Tumor suppressor-3 Page 6 of 7 MRC-Holland ® MLPA Description version 04; 05-02-2010 134.80 70000 177.46 60000 291.22 96.77 50000 201.14 91.52 228.52 256.24 40000 105.77 336.92 474.10 309.08 389.86 30000 100.97 418.44 86.08 443.72 364.17 20000 10000 106.73 Dye Signal 61.64 0 100 150 200 250 300 350 400 450 500 Size (nt) Figure 2. Capillary electrophoresis pattern from a sample of approximately 50 ng digested human male control DNA analyzed with SALSA MLPA kit ME003-A1 Tumor suppressor-3 (lot 0608). Implemented Changes – the following has been altered compared to the previous product description version(s). Version 04 (05) - ‘Basic research’ note added on page 1 - Columns in Table 2 have been adjusted. Textual changes on page 1 and 2, tables have been numbered. SALSA MLPA kit ME003 Tumor suppressor-3 Page 7 of 7