* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Organization of Genes Differs in Prokaryotic and Eukaryotic DNA
Ridge (biology) wikipedia , lookup
Oncogenomics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Protein moonlighting wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA interference wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Human genome wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA silencing wikipedia , lookup
Designer baby wikipedia , lookup
Minimal genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Frameshift mutation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome evolution wikipedia , lookup
History of RNA biology wikipedia , lookup
Polyadenylation wikipedia , lookup
Microevolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Helitron (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Messenger RNA wikipedia , lookup
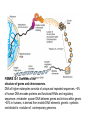
Organization of Genes Differs in Prokaryotic and Eukaryotic DNA Chapter 10 p.110-114 Arrangement of information in DNA----- requirements for RNA Common arrangement of protein-coding genes in prokaryotes= e.g Operon ----------------- operates as a unit from a single promoter. Transcription of operon produces a continuous strand of mRNA: carries message for a related series of proteins Genes in prokaryotic packed with very few noncoding gaps---- DNA transcribed into colinear mRNA-----translated into protein. Economic clustering of genes devoted to a single metabolic function In Prok. does not occur in eukaryotes Yeasts= metabolically similar to bacteria. Eukaryotic genes in multicellular devoted to a single pathway physically separated in DNA Exons and Introns (Introns rare in Bacteria and Archea,uncommon in unicellular eukaryotes e.g baker’s yeast Introns present in DNA of viruses infect eukaryotic cells (Introns first discovered in viruses) Genes located on different chromosomes, transcribed from its own promoter producing one mRNA, translated to a single polypeptide Eukaryotic Precursor mRNAs Are Processed to Form Functional mRNAs Transcription & translation occur concurrently in prokaryotes but not in Eukaryotic, Why? Primary transcripts = precursor mRNAs (pre-mRNAs) undergo modifications at both ends &RNA processing, to yield a functional mRNA ------- mRNA exported to cytoplasm 5_ end of a nascent RNA chain emerges from surface of RNA polymerase II ---------- acted on by enzymes that synthesize the 5_ cap=7-methylguanylate connected to terminal nucleotide of RNA by an unusual 5_,5_ triphosphate linkage The cap function? -Protection from enzymatic action -Export to cytoplasm -bound by a protein factor required to begin translation Processing at 3_ end of a pre-mRNA involves cleavage by an endonuclease----free 3_-hydroxyl group to adenylic acid residues added one at a time by poly(A) polymerase -----poly(A) tail ( 100–250 bases) shorter in yeasts invertebrates than in vertebrates ----poly A addition doesnt not require a template . Final step in processing of mRNA is RNA splicing: e.g Globin gene mRNAs produced by RNA processing retain noncoding regions: 5_ and 3_ untranslated regions (UTRs). 5_ UTR hundred or more nucleotides / 3_ UTR several kilobases in length . Prokaryotic mRNAs usually have 5_ and 3_ UTRs shorter than eukaryotic mRNAs Alternative RNA Splicing Increases the Number of Proteins Expressed from a Single Eukaryotic Gene Proteins in Euk. have a multidomain tertiary structure Repeated protein domains encoded by one exon/ small number of exons that code for identical or nearly identical amino acid sequences Multiple introns in eukaryotic genes permits expression of multiple, related proteins (isoforms/different forms) from a single gene by alternative splicing. e.g Fibronectin (multidomain extracellular adhesive protein) Fibroblasts produce fibronectin mRNAs contain exons EIIIA&EIIIB ------------ encode a.a bind tightly to proteins in fibroblast plasma membrane. Alternative splicing of fibronectin primary transcript in hepatocytes, lack EIIIA & EIIIB exons-----does not adhere tightly to fibroblasts ----- circulate in blood. formation of blood clots , domains of hepatocyte fibronectin binds to fibrin, one of the principal constituents of clots fibronectin interacts with integrins on the membranes---- activated platelets ----- expanding clot by addition of platelets. ---------------------------------------------------------------------------Sequencing of genomic DNA revealed 60% of all human genes expressed as alternatively spliced mRNAs ▲ FIGURE 4-14 Overview of RNA processing to produce functional mRNA in eukaryotes. ▲ FIGURE 4-13 Structure of the 5 methylated cap of eukaryotic mRNA. ▲ FIGURE 4-15 Cell type–specific splicing of fibronectin pre-mRNA in fibroblasts and hepatocytes. The ≈75-kb fibronectin gene (top) contains multiple exons. The EIIIB and EIIIA exons (green) encode binding domains for specific proteins on the surface of fibroblasts. Whereas exons are spliced out of fibronectin mRNA in hepatocytes. MOLECULAR STRUCTURE OF GENES AND CHROMOSOMES p.405-408 P408-424 By beginning of 21st century, completed sequencing, entire genomes of viruses, bacteria & budding yeast S. cerevisiae, D. melanogaster, and humans - sequencing data revealed large portion of genomes of higher eukaryotes -----more 95% human chromosomal DNA non-coding -regions similar but not identical( application: e.g DNA “fingerprint”) -Some repetitious DNA sequences not found in constant positions in the same species--------Mobile” DNA elements (in prokaryotic &eukaryotic) Mobile DNA-----can cause mutations when move to new sites in genome. ------have no function in life cycle, probably played role in evolution. In higher eukaryotes, DNA regions encoding proteins— genes nonfunctional DNA=noncoding introns common within genes Sequencing of same protein-coding gene (Exons)in a variety of eukaryotic species Shown relatively similar sequences sequence variation, including total loss, occurs among introns(less functional significance) Objectives: - a molecular definition of genes - complexities that arise in higher organisms from processing of mRNA precursors into alternatively spliced mRNAs. - special properties of mobile DNA. - packaging of DNA and proteins into compact complexes, large-scale structure of chromosomes, and e functional elements required for chromosome duplication and segregation. Molecular Definition of a Gene A gene= entire nucleic acid sequence necessary for synthesis of a functional gene product (polypeptide or RNA). A gene also includes: - all DNA sequences required for synthesis of RNA transcript e.g transcription-control regions= enhancers lie 50 kb or more from coding region. - noncoding regions= sequences specify 3_ cleavage & polyadenylation( poly(A) sites) - splice sites 10.1 -most genes transcribed into mRNAs, and some transcribed into tRNAs & rRNAs =tRNA, rRNA genes?? Most Eukaryotic Genes Produce Monocistronic mRNAs and Contain Lengthy Introns -many bacterial mRNAs polycistronic---------- a single mRNA molecule (e.g., mRNA encoded by trp operon) includes coding region(Cistrones) for several proteins , function together in a biological process. bacterial polycistronic mRNA a ribosome binding site located near start site for each cistrons------- Translation initiation begin at any of these multiple internal sites ----------------producing multiple proteins -Most eukaryotic mRNAs monocistronic------- each mRNA molecule encodes a single protein. In most eukaryotic mRNAs, 5_-cap directs ribosome binding, and translation begins at closest AUG start codon---------- translated to give a single type of polypeptide Simple and Complex Transcription Units Are Found in Eukaryotic Genomes Cluster of genes that form a bacterial operon= a single transcription unit transcribed from a promoter into a single primary transcript. Eukaryotic transcription units classified into two types, depending on fate of primary transcript. -1-------Primary transcript produced from a simple transcription unit processed to yield a single type of mRNA---encoding a single protein. Mutations in exons, introns and transcription-control regions ---influence expression of protein 2- complex transcription units---- common in multicellular organisms, ----- primary RNA transcript processed more than one way----Leading to formation of mRNAs containing different exons . Each mRNA monocistronic----translated into a single polypeptide, Multiple mRNAs can arise from a primary transcript in three ways: 1. exon skipping :Use different splice sites, producing mRNAs with the same 5_ and 3_ exons but different internal exons. . 2. Use of alternative poly(A) sites, producing mRNAs that share same 5_ exons but have different 3_ exons. 3. Use of alternative promoters, producing mRNAs -----have different 5_ exons and common 3_ exons. A gene expressed selectively in two or more types of cells transcribed from distinct cell-type-specific promoters Examples of all three types of alternative RNA processing occur during sexual differentiation in Drosophila Differences in RNA splicing of primary fibronectin transcript in fibroblasts and hepatocytes determines whether or not the secreted protein includes domains that adhere to cell surfaces (see Figure 4-15). The relationship between a mutation and a gene not straightforward in complex transcription units. A mutation in control region or in an exon shared by alternative mRNAs will affect alternative proteins encoded by complex transcription unit. Mutations in an exon present in one of alternative mRNAs will affect only the protein encoded by that mRNA. Genetic complementation tests : To determine if two mutations in same or different genes In complex transcription unit, mutations d and e complement each other in genetic complementation test, even they occur in same gene-----a chromosome with mutation d can express a normal protein encoded by mRNA2 and a chromosome with mutation e can express a normal protein encoded by mRNA1 .! However, a chromosome with mutation c in an exon common to both mRNAs not complement either mutation d or e. In other words, mutation c would be in the same complementation groups as mutations d and e, even though d and e themselves would not be in the same complementation group FIGURE 10-1 Overview of the structure of genes and chromosomes. DNA of higher eukaryotes consists of unique and repeated sequences. ~5% of human DNA encodes proteins and functional RNAs and regulatory sequences ;remainder spacer DNA between genes and introns within genes. ~50% in humans, is derived from mobile DNA elements, genetic symbiots contributed to evolution of contemporary genomes. FIGURE 10-2 : A simple transcription unit includes a region that encodes one protein, Introns lie between exons (blue rectangles) removed during processing of primary transcripts (dashed red lines); ---------not in functional monocistronic mRNA. Mutations in a transcription-control region (a, b) may reduce or prevent transcription, ------------- reducing or eliminating synthesis of the encoded protein. A mutation within exon (c) may result in abnormal protein with diminished activity. A mutation within an intron (d ) ----- introduces a new splice site----- in abnormally spliced mRNA -------- nonfunctional protein. (b) Complex transcription units produce primary transcripts processed in alternative ways. (Top) a primary transcript contains alternative splice sites----- processed into mRNAs with same 5 and 3 exons but different internal exons. (Middle primary transcript has 2 poly(A) sites-------processed into mRNAs with alternative 3 exons. (Bottom) alternative promoters (f or g) active in different cell types, mRNA1, produced in a cell type , f activated, has a different exon (1A) than mRNA2 , produced in a cell type ,g activated .Mutations in control regions (a and b) and c within exons shared by alternative mRNAs affect the proteins encoded by both alternatively processed mRNAs. In contrast, mutations (designated d and e) within exons unique to one of alternatively processed mRNAs affect only protein translated from that mRNA. For genes transcribed from different promoters in different cell types (bottom), mutations in different control regions (f and g) affect expression