* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Controlling the Code: molecules at work
Site-specific recombinase technology wikipedia , lookup
Dominance (genetics) wikipedia , lookup
DNA supercoil wikipedia , lookup
Epigenomics wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
RNA interference wikipedia , lookup
Y chromosome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Designer baby wikipedia , lookup
Messenger RNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Neocentromere wikipedia , lookup
Point mutation wikipedia , lookup
Microevolution wikipedia , lookup
RNA silencing wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of RNA biology wikipedia , lookup
Epitranscriptome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Non-coding RNA wikipedia , lookup
X-inactivation wikipedia , lookup
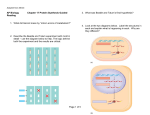
Controlling the Code: molecules at work The lac operon 2. (a) Write a description of the role of the operator in B-gal production. • What is the operator? The operator is a segment of DNA located between the promoter and the enzyme genes. It acts as a switch by determining whether RNA polymerase can attach to the promoter and start transcribing. • When is the repressor protein bound to the operator? The repressor is bound to the operator when there is no lactose present. • When does transcription occur? Why? Transcription occurs when the repressor protein falls off the operator because it no longer blocks RNA polymerase from binding with the promoter. • What is the role of lactose in the initiation of transcription? Lactose binds to the repressor protein. This causes the repressor protein to be released from the operator site. RNA polymerase can then bind to the promoter. • How does feedback (both negative and positive) regulate the production of B-gal? The lac operon is a model of negative regulation because an inhibitor must be removed from the DNA to turn on the gene. It is a model of positive regulation because, as well as getting rid of the inhibitor, an activator must attach to the DNA to turn on B-gal synthesis. Molecules for hire 2. Circle the number of each Work Wanted ad that accurately describes a part of the regulation of gene transcription associated with the production of B-gal. Work Wanted Ads: Seeking Operon Position Ad #1011, Seeking Repressor Position Ad #1111, Experienced Promoter Seeks Position Ad #1122 3. Complete the Work Wanted posting for mRNA. Be sure to include its role in B-gal production and “who” it associates with during the process. Add the information to the Work Wanted ad sheet. Messenger RNA Seeks Steady Work Ad #1133 Student answers will vary but should include that RNA is transcribed when lactose is present. It is then that the repressor is released from the operator and no longer blocks the attachment of RNA polymerase to the promoter. This allows transcription to begin. 1 Further exploration Explanation Explain how the tortoiseshell pattern observed on some female cats provides evidence that packing plays an important role in gene regulation. Highly compacted chromatin can be found in some regions of interphase chromosomes. These regions are typically not expressed. Taking this a step further, one X chromosome in female mammals is highly compacted and almost entirely inactive. This X chromosome inactivation occurs during early embryonic development. One of the two X chromosomes in each cell is inactivated at random. The inactivation is inherited by all of a cell’s descendents. The tortoiseshell gene in cats is located on the X chromosome. The tortoiseshell genotype requires the presence of two different alleles—orange fur and nonorange (black) fur. Orange patches of fur are formed by populations of cells in which the X chromosome with the orange allele is active (not compacted). Black patches of fur form where the X chromosome containing the nonorange (black) allele is active. 2