* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Peptide bonds and side chains Peptide bonds
Survey
Document related concepts
Ancestral sequence reconstruction wikipedia , lookup
Point mutation wikipedia , lookup
Interactome wikipedia , lookup
Biosynthesis wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Biochemistry wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Homology modeling wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Peptide synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Transcript
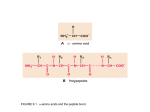
Protein physics, Lecture 5 Peptide bonds: resonance structure Properties of proteins: Peptide bonds and side chains Proteins are linear polymers However, the peptide binds and side chains restrict conformational possibilities How do the peptide backbone and the amino acid side chains influence protein structure? Planar Peptide bond Peptide bonds are ‘stiff’ Planarity of the peptide bonds is due to: • Partial double bond character • Large dipole moment that inhibits rotation Resonance forms of a typical peptide group. The uncharged, single-bonded form (typically ~60%) is shown on the left, whereas the charged, double-bonded form (typically ~40%) is on the right. Dihedral angles Rotation around a bond Planarity of bond means that ‘cis’ and ‘trans’ forms are possible Trans form is much more common due to less steric hindrance Cis form is most common in Proline residues Compare with bond angle Dihedral angles Dihedral angles CE In general there are 4 dihedral angles involved in describing protein structure F Side chain \ CD CE F \ CD N N I Z C Z O I C Backbone Peptide bond Planar O I and \ are flexible And account for freedom One amino acid of protein structure Carbonyl group Z= 0 or 180 Protein conformation can be described in terms of the amino acid sequence and dihedral angles: Ii, \i, Fi for each amino acid residue Flexibilty of the polypetide chain is restricted by • Stiff peptide bond • Steric hindrance – (ie avoiding overlap of atoms) Ramachandran et al GN Ramachandran (1922-2001) Ramachandran plot for glycine • G.N. Ramachandran (1963) used computer models of small polypeptides to systematically vary and with the objective of finding stable conformations • For each conformation, the structure was examined for close contacts between atoms • Atoms were treated as hard spheres with dimensions corresponding to their van der Waals radii • Therefore, and angles which cause spheres to collide correspond to sterically disallowed conformations of the polypeptide backbone Ramachandran plot • Allowed regions are in the 4 corners of the plot • Prohibitive contacts are indicated • Note that glycine has no side chain and is therefore the most flexible •Plot of vs. •The computed angles which are sterically allowed fall on certain regions of plot Can improve using potential energy function rather than hard sphere model. Just the crosshatched regions Ramachandran plot for with CE Ramachandran plot for with CE Allowed regions correspond to angles that yield E sheet D helix Left hand helix Repeating values of and along the chain result in regular structure Experimental Ramachandran plot , distribution from over 80,000 AA residues from high-resolution protein structures (x-ray crystallography) Prediction using potential energy function • About 5% of observed conformations fall in ‘forbidden regions’ • Flexibility of peptide bond needs to be taken into account to improve this • Deviations of 5o bond angle; 0.05Å bond length or 12o torsion angle (Z) increases the potential energy by about 1/kcal/mol each Protein structure example The structure of cytochrome C shows many segments of helix and the Ramachandran plot shows a tight grouping of , angles near -50,-50 alpha-helix cytochrome C Ramachandran plot Protein structure example Similarly, repetitive values in the region of = -110 to –140 and = +110 to +135 give beta sheets. The structure of plastocyanin is composed mostly of beta sheets; the Ramachandran plot shows values in the –110, +130 region: Side chain properties How do the side chains influence protein structure? Glycine • Side chain is just H • Increases side chain flexibility (see Ramachandran plot) • Can fit chain into small spaces • Frequency restricted – too much would render chains to flexible and lose 3D shape Alanine • Has one methyl group as side chain • Smallish – nonpolar • No real preference for inside or surface of protein beta-sheet • Very abundant (due to simplicity and availability?) plastocyanin Ramachandran plot Side chain properties Side chain properties Polar side chains Branched side chains are stiffer • Cys, Ser, Thr, Asn, Gln, Tyr • Can form hydrogen bonds • Often on surface of protein • Val, Ile, Leu • Reduce possibilities for chain folding Cysteine • Has unique property: only side chain with exposed S atom Aromatic residues • Phe, Trp, Tyr, His • Can form disulfide bond with another • Contain one methylene group ’spacer’ Cys further along the chain • Without this severe steric hinderance • Used to maintain a given structure of the protein or to respond to oxidation • Would make chain too stiff •Intrinsically fluorescent – particularly Trp Large non-polar residues • Leu, Ile, Phe, Pro, Trp • Predominantly found in the interior of the molecule Disulfide bond Side chain properties Side chain properties Negative charged side chains Proline • Back bone is part of the cyclic group • Backbone section has a bend • Invokes bends in the protein chain or kinks in helices • Asp, Glu • Negative charge at physiological pH (lose an H+) • Usually found on protein surface Positive charged side chains • Lys, Arg Histidine • Positive charge at physiological pH (gain an H+) • Side chain has pK value of 6.0 • Can be charged or uncharged in the physiological pH range • Two readily available states – useful as a catalyst • Involved in the active site of most enzymes • Usually found on protein surface Hydrophobicity • Folding process of polypetide chain depends on hydrophobicity (non-polarity) of side chains • Formation of hydrophobic core is an essential driving force in protein folding