* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 1. Protein Interactions
Structural alignment wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein design wikipedia , lookup
Homology modeling wikipedia , lookup
Implicit solvation wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Alpha helix wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
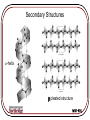
Protein Interactions with Biomaterials Topics: •Thermodynamics of Protein Adsorption •Protein Structure •Protein Transport and Adsorption Kinetics MSE-536 Thermodynamics For a reaction to spontaneously occur, the change in Gibbs free energy, DG, must be <0: DG DH TDS DGads DG prot DGsol DGSurf G = Gibbs free energy H = enthalpy (energy available to do work) S = entropy (disorder) MSE-536 Thermodynamics of Protein Adsorption Hydrophobicity: Hydrophobic areas attract hydrophobic areas Charge: Opposite charges attract Size: Larger molecules have more active sites Structure: the stability (strength of intramolecular bonds) and molecule unfolding rate MSE-536 Surface features and their interactions with proteins: • Topography: greater texture means greater interaction • Composition: Chemistry governs types of interactions • Hydrophobicity: hydrophobic surfaces bind more protein • Heterogeneity: non-uniform surfaces have many different types of domains to interact with proteins • Potential: surface charge affects charge distributions of ions in solution and proteins MSE-536 Protein Structure Proteins are polymeric chains of amino acids. Each of the 20 standard amino acids have a oneletter symbol. A sequence of three symbols, as shown for RNA (right) is called a Amino acids have a central codon carbon atom attached to a hydrogen, a carboxyl group (COOH) and an amine group (NH2) MSE-536 The pK value is related to the pH of the amino acid. Higher values are more acidic (lower pH) MSE-536 MSE-536 Proteins (polypeptides) are formed from condensation reactions between amino acids (peptide bonds). MSE-536 Secondary Structures a-helix b pleated structure MSE-536 Tertiary and Quaternary Structures Interactions between side chains control how the protein folds in three and four dimensions. These interactions include: •Covalent bonding •Ionic interactions •Hydrogen bonding •Hydrophobic interactions MSE-536 Protein Transport and Adsorption Kinetics Four main types of protein transport: 1. Diffusion 2. Thermal convection 3. Flow (convective transport) 4. Coupled transport (combinations of 1-3) A concentration gradient drives diffusion, while a temperature gradient creates thermal convection MSE-536 Diffusion is Fick’s 2nd law, with the addition of a contribution from flow: dC dC 1 C V D r dt dz r r r The velocity profile is given by: 2 2Q r V 1 2 R R V = velocity = viscosity Here in cylindrical coordinates C = concentration D = diffusivity Q = volumetric flow rate MSE-536 Initial absorption rate is high on a clean surface Rate slows as surface becomes covered Further absorption occurs as molecules rearrange to create new free surface MSE-536 Protein exchange on a material surface. The initial protein (light gray) is wedged out of the way by the newer proteins (dark gray), which have a greater affinity for the material MSE-536