* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Part 2
Bottromycin wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Expanded genetic code wikipedia , lookup
Genetic code wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Gene expression wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Biochemistry wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Circular dichroism wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein domain wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Protein folding wikipedia , lookup
Interactome wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein structure prediction wikipedia , lookup
Metalloprotein wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Phosphorylation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein purification wikipedia , lookup
Protein adsorption wikipedia , lookup
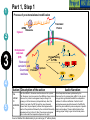
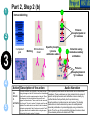
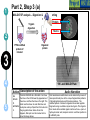
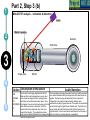
Post-translational modifications a Many proteins undergo chemical modifications at certain amino acid residues following translation. These modifications are essential for normal functioning of the protein and are carried out by one or more enzyme catalyzed reactions. Harini Chandra Master Layout (Part 1) 1 Commonly observed PTMs This animation consists of 4 parts: Part 1: Overview of PTMs Part 2: Gel-based detection techniques for PTMs Part 3: MS-based detection techniques for PTMs Part 4: Microarray-based detection techniques for PTMs Phosphorylation Glycosylation Acylation 2 Alkylation Hydroxylation 3 Cytosol Endoplasmic reticulum (ER) P P Signal sequence 4 Protein translation PTMs Cleaved protein CH3 CH3 Glc Glc 5 Source: Modified from Biochemistry by A.L.Lehninger, 4th edition (ebook) 1 2 Definitions of the components: Part 1 – Overview of PTMs 1. Post-translational modification (PTM): The chemical modifications that take place at certain amino acid residues after the protein is synthesized by translation are known as posttranslational modifications. These are essential for normal functioning of the protein. Some of the most commonly observed PTMs include: a) Phosphorylation: The process by which a phosphate group is attached to certain amino acid side chains in the protein, most commonly serine, threonine and tyrosine. 3 b) Glycosylation: The attachment of sugar moieties to nitrogen or oxygen atoms present in the side chains of amino acids like aspargine, serine or threonine. c) Acylation: The process by which an acyl group is linked to the side chain of amino acids like aspargine, glutamine or lysine. 4 d) Alkylation: Addition of alkyl groups, most commonly a methyl group to amino acids such as lysine or arginine. Other longer chain alkyl groups may also be attached in some cases. e) Hydroxylation: This PTM is most often found on proline and lysine residues which make up the collagen tissue. It enables crosslinking and therefore strengthening of the muscle fibres. 5 1 2 Definitions of the components: Part 1 – Overview of PTMs 2. Protein translation: The process by which the mRNA template is read by ribosomes to synthesize the corresponding protein molecule on the basis of the three letter codons, which code for specific amino acids. 3. Cytosol: A cellular compartment that serves as the site for protein synthesis. 4. Signal sequence: A sequence that helps in directing the newly synthesized polypeptide chain to its appropriate intracellular organelle. This sequence is most often cleaved following protein folding and PTM. 3 5. Endoplasmic reticulum: A membrane-bound cellular organelle that acts as a site for posttranslational modification of the newly synthesized polypeptide chains. 6. Cleaved protein: The protein product obtained after removal of certain amino acid sequences such as N- or C-terminal sequences, signal sequence etc. 4 5 1 Part 1, Step 1 Process of post-translational modification Translated Protein mRNA Ribosome Cytosol 2 P 3 Endoplasmic reticulum (ER) Removal of certain N- and Cleaved protein C-terminal Protease residues P Protein folding & PTMs CH3 CH3 Glc Glc 4 Action Description of the action 5 As shown in animatio n. First the ‘mRNA’ & ‘ribosome’ must be shown in the ‘cytosol’. The ‘ribosome’ must move across the mRNA as shown and as it moves, the ‘protein’ must appear slowly as though it is growing out of the ribosome (not depicted here). Next, this ‘protein’ must enter the ‘ER’ through the green channels shown. Next, the pie-shaped ‘protease’ must appear which must cut the pink strand followed by the red strand followed by appearance of text on the left. Next, the arrows must appear one at a time with their respective figures on the right. Audio Narration Once the protein has been synthesized by the ribosome from its corresponding mRNA in the cytosol, many proteins get directed towards the endoplasmic reticulum for further modification. Certain N and C terminal sequences are often cleaved in the ER after which they are modified by various enzymes at specific amino acid residues. These modified proteins then undergo proper folding to give the functional protein. Source: Modified from Biochemistry by A.L.Lehninger, 4th edition (ebook) Part 1, Step 2 1 Different types of PTMs & their modification sites Ser, Thr, Tyr Pro, Lys 2 Phosphorylation Asn, Glycosylation Ser, Thr Lys, Arg 3 Acylation Alkylation Hydroxylation Asn, Gln, Lys 4 Action Description of the action 5 As shown in animatio n. First show the pie chart as depicted. Next, each segment must be highlighted sequentially along with appearance of the corresponding text in the boxes as depicted. Audio Narration There are several types of post translational modifications that can take place at different amino acid residues. The most commonly observed PTMs include phosphorylation, glycosylation, methylation as well as hydroxylation and acylation. Many of these modifications, particularly phosphorylation, serve as regulatory mechanisms for protein action. 1 Part 1, Step 3 Increased complexity of proteome due to PTMs A A C G G U G C C G U G C A C G C A C U A C G C A C U Expected protein structure 2 Gene sequence Actual protein structure 3 Glc P CH3 4 Action Description of the action 5 As shown in animatio n. First show the ‘gene sequence’ on top followed by the arrow to the left and the blue structure. This must be zoomed into to show the inset below. The red cross must then appear on this arrow. Next, the arrow to the right must appear followed by the pink structure which must again be zoomed into to show the inset below. Audio Narration The final structure of functional proteins most often does not correlate directly with the corresponding gene sequence. This is due to the PTMs that occur at various amino acid residues in the protein, which cause changes in interactions between the amino acid side chains thereby modifying the protein structure. This further increases the complexity of the proteome as compared to the genome. 1 Part 1, Step 4 Phosphorylation reactions COO- COOKinase 2 C H NH3+ 3 R H OH ATP Amino acid residue ADP CH2 NH3+ C R O PO43- Phosphorylated residue Ser CH Thr R CH3 Tyr CH2 4 Action Description of the action 5 As shown in animatio n. First show the figure on the top left entering followed by the box below having the various three arrows from “R”. Next the arrow must ease in along with the curved arrow below. Finally, the figure on the right must appear. Audio Narration Phosphorylation of amino acid residues is carried out by a class of enzymes known as kinases that most commonly modify side chains of amino acids containing a hydroxyl group. Phosphorylation requires the presence of a phosphate donor molecule such as ATP, GTP or other phoshorylated substrates. Serine is the most commonly phosphorylated residue followed by threonine and tyrosine. Removal of phosphate groups is carried out by the phosphatase enzyme and thus this forms one of the most important mechanisms for regulation of proteins. th 1 Part 1, Step 5 Glycosylation reactions N-linked Glycosylation COO- 2 H Glycosyl transferase C CH2 H C CH2 CONH2 CON NH3+ NH3+ Asn 3 Sugar residues COO- N-linked amino acid O-linked Glycosylation COO- COOGlycosyl transferase H C R C H R O OH NH3+ 4 NH3+ Ser/Thr O-linked amino acid Action Description of the action 5 As shown in animatio n. First show appearance of the figure on top let with heading followed by arrow and finally product on right. Similar animation must be carried out for the second reactions. Audio Narration Glycosylation involves the enzymatic addition of saccharide molecules to amino acid side chains. This can be of two types – N-linked glycosylation, which links sugar residues to the amide group of aspargine and O-linked glycosylation, which links the sugar moieties to the hydroxyl groups of serine or threonine. Suitable glycosyl transferase enzymes catalyze these reactions. Sugar residues that are attached most commonly include galactose, mannose, glucose, Nacetylglucosamine, N-acetylgalactosamie as well as fucose. th Master Layout (Part 2) 1 This animation consists of 4 parts: Part 1: Overview of PTMs Part 2: Gel-based detection techniques for PTMs Part 3: MS-based detection techniques for PTMs Part 4: Microarray-based detection techniques for PTMs Gel scanning Gel staining 2 3 Specific probe antibodies 4 Electrophoresis 1. Pro-Q-diamond Blotting SDS-PAGE/ 2-DE gel 5 Detection Nitrocellulose sheet 2. Immunoblotting Labeled secondary Abs 1 2 Definitions of the components: Part 2 – Gel-based detection techniques for PTMs 1. Pro-Q-diamond: This fluorescent dye is capable of detecting modified proteins that have been phosphorylated at their serine, threonine or tyrosine residues. They are suitable for use with electrophoretic techniques or with protein microarrays and offer sensitivity down to few ng levels, depending upon the format in which they are used. This dye can also be combined with other staining procedures thereby allowing more than one detection protocol on a single gel. a) Gel staining: The process by which the protein bands on an electrophoresis gel are stained by suitable dyes for visualization. 3 4 5 b) Gel scanning: The visualization of the stained protein bands on an electrophoresis gel by exciting it at a suitable maximum wavelength such that the dye absorbs the light and emits its own characteristic light at another emission wavelength. 2. Immunoblotting: This process, also known as Western blotting, is a commonly used analytical technique for detection of specific proteins in a given mixture by means of specific antibodies to the given target protein. a) Electrophoresis: Electrophoresis is a gel-based analytical technique that is used for separation and visualization of biomolecules like DNA, RNA and proteins based on their fragment lengths or charge-to-mass ratios using an electric field. The protein mixture is first separated by means of a suitable electrophoresis technique such as SDS-PAGE or Twodimensional Electrophoresis. 1 2 Definitions of the components: Part 2 – Gel-based detection techniques for PTMs b) Blotting: The process by which the proteins separated on the electrophoresis gel are transferred on to another surface such as nitrocellulose by placing them in contact with each other. c) Nitrocellulose sheet: A membrane or sheet made of nitrocellulose onto which the protein bands separated by electrophoresis are transferred for further probing and analysis. 3 4 5 d) Specific probe antibodies: Antibodies that are specific to a particular protein modification can be used as probes to detect those proteins containing that particular PTM. Protein phosphorylation is commonly detected using anti-phosphoserine, phosphothreonine or phosphotyrosine antibodies. Recently, specific motif antibodies have also been developed which detect a particular sequence of motif of the protein that contains a PTM. e) Labeled secondary Abs: Antibodies labeled with a suitable fluorescent dye molecule are used to detect the interaction between the modified protein and its antibody by binding to another domain of the probe antibody. 1 Part 2, Step 1 (a) Pro-Q-diamond staining Dye stains the get Protein bands Excess dye phosphorylated fixed on gel and removed protein bands only. minimize diffusion. Completed 2-DE gel 2 Tubing connected & outlet opened 3 Tray with fixing solution Pro-Q-diamond stain Washing solution (methanol + acetic acid) (methanol + acetic acid) 4 Action Description of the action 5 As shown in animatio n. First show appearance of the ‘2-DE’ gel on top along with tray having light grey solution. Gel must be placed horizontally in the tray after which the dialogue box must appear. Next the solution must be drained out as shown and then re-filled with the red solution. Again the dialogue box must appear after which the solution is drained out and the tray re-filled with light blue solution. The last dialogue box then appears and this solution is then drained out. Audio Narration Protein phosphorylation can be detected using a novel gel-based detection technique. Proteins separated on a 2-DE gel are first placed in a fixing solution containing methanol and acetic acid which fixes the protein bands on to the gel and minimizes any diffusion. They are then stained using the Pro-Q-diamond staining solution which selectively stains only phosphoproteins on the gel. The excess stain is then washed off with a solution of methanol and acetic acid. 1 Part 2, Step 1 (b) Gel scanning Gel scanner 2 Stained gel Emission maxima – 580 nm Decreasing molecular weight 3 Phosphoprotein image Decreasing pH 4 Action Description of the action 5 Gel removed from scanner As shown in animatio n. PLEASE REDRAW ALL IMAGES. First show the ‘scanner’ and the ‘stained gel’ image. The lid of the scanner must be opened and the gel must be placed horizontally in it & lid must be closed. Next the yellow beam must appear followed by the image below and all the labels. Next the scanner lid must be opened and the gel must be removed from it. Audio Narration The stained gel is then scanned at its excitation wavelength using a gel scanner. The gel image obtained shows the protein bands corresponding to only the phosphoproteins present. This image is saved and the gel is then removed from the scanner for treatment with the second stain, a procedure known as dual staining. 1 Part 2, Step 1 (c) Dual staining with SYPRO-Ruby Red Dye stains all Excess dye protein bands. removed Tubing connected & outlet opened 2 3 Washing solution SYPRO-Ruby red (methanol acetic acid) staining+ solution 4 Action Description of the action 5 As shown in animatio n. The gel from the previous slide, after being removed from the scanner is placed in a tray containing the red ‘SYPRO-Ruby Red’ stain as shown. The dialogue box must appear after which the tubing must appear and the solution must be drained. The light blue solution must then be added to the tray and the dialogue box on top must appear after which the solution must again be drained as depicted. Audio Narration The scanned gel is then removed from the scanner and placed in the SYPRO-Ruby Red fluorescent dye solution. This dye stains all the protein spots present on the gel thereby providing a total protein image with sensitivity down to nanogram level. Excess dye is then washed off using a solution of methanol and acetic acid. Fluorescence Gel scanning Gel scanner 2 Phosphoprotein image Emission maxima – 610 nm Total protein image Decreasing molecular weight 3 Decreasing pH 4 A comparative profile between total protein image and phosphoprotein image enables detection of phosphorylated proteins. Action Description of the action 5 Phosphoprotein image Stained gel As shown in animatio n. PLEASE REDRAW ALL IMAGES. First show the ‘scanner’ and the ‘stained gel’ image. The lid of the scanner must be opened and the gel must be placed horizontally in it & lid must be closed. Next the red beam must appear followed by the image below and all the labels. The graphs along with the ‘phosphoprotein image’ must then appear followed by the green text box. Fluorescence 1 Part 2, Step 1 (d) Total protein image by SYPRO-Ruby Red Audio Narration The gel stained with SYPRO-Ruby Red is then scanned in the gel scanner at its excitation maxima. The image produced will have more number of spots since all proteins present on the gel are detected. This dual staining procedure provides a useful comparative profile of the phosphoproteins and the total proteins on the gel, thereby enabling detection of the phosphorylated proteins. 1 Part 2, Step 2 (a) Proteins focused on IPG strip Immunoblotting SDS-PAGE - 2-D Electrophoresis Sample loading Cathode 2 3 Direction of migration Protein mixture Acrylamide gel Direction of migration + Anode Buffer Completed stained gels 4 Action Description of the action 5 As shown in animatio n. Audio Narration PLEASE REDRAW ALL IMAGES. First show the setup on the Protein mixture containing phosphorylated as left and right with their labels. Next the hand must appear on well as other unmodified proteins can be the left and the ‘strip’ on the right. The hand must move into separated by a suitable electrophoresis technique. SDS-PAGE and two dimensional one of the wells and the thick blue band must appear. The strip on the right must be placed on the light blue surface as gel electrophoresis are most commonly used shown. Next, the downward arrows must appear along with for protein separation. These separated text followed by the blue bands on the left and the spots on proteins on the gel are used for further analysis. the right. Finally the images below must appear. 1 Part 2, Step 2 (b) Immunoblotting Proteins phosphorylated at Tyr residues 2 Completed gels Nitrocellulose Blotting sheet 3 4 5 Specific phosphotyrosine antibodies added Detection using labeled secondary antibodies Proteins phosphorylated at Tyr residues Action Description of the action As shown in animatio n. Audio Narration PLEASE REDRAW ALL IMAGES. First show the two The separated protein bands are then blotted onto a nitrocellulose blue gel images on the left. Next show the ‘nitrocellulose membrane. These membranes are then probed either by means of sheet’ which must be superimposed on the gel. When specific anti-phospho-amino acid antibodies or more recently, by this happens, the blue bands and blue spots must motif antibodies that specifically bind to proteins having appear on these sheets. These must then be removed phosphorylation at a particular amino acid residue. This binding from the gel. The pink inverted Y objects must then be interaction can then be detected by means of suitably labeled added to the sheets which must bind to the blue bands and spots as depicted. Next, the brown inverted Y with secondary antibodies or by autoradiography using a radioactive the green label must be added which must glow upon probe. Thus, the use of immunoblotting technique has been shown to be extremely effective for detection of PTMs. contact with the pink inverted Y objects. Master Layout (Part 3) 1 This animation consists of 4 parts: Part 1: Overview of PTMs Part 2: Gel-based detection techniques for PTMs Part 3: MS-based detection techniques for PTMs Part 4: Microarray-based detection techniques for PTMs MALDI-TOF-MS 2 Ion source Flight tube Detector 3 1. MALDI-TOF analysis Affinity columns 4 Liquid chromatography 5 Tandem MS 2. LC-MS/MS approach 1 2 3 4 Definitions of the components: Part 3 – MS-based detection techniques for PTMs 1. MALDI-TOF-MS: A mass spectrometry instrument that produces charged molecular species in vacuum, separates them by means of electric and magnetic fields and measures the mass-to-charge ratios and relative abundances of the ions thus produced. It has the following components: a) Ion source: The ion or ionization source is responsible for converting analyte molecules into gas phase ions in vacuum. The technology that enables this is termed soft ionization for its ability to ionize non-volatile biomolecules while ensuring minimal fragmentation and thus, easier interpretation. In MALDI-TOF-MS, the ion source used is MALDI, in which the target analyte is embedded in dried matrix-sample and exposed to short, intense pulses from a UV laser. b) Flight tube: Connecting tube between the ion source and detector within which the ions of different size and charge migrate to reach the detector. The Time-of-Flight mass analyzer correlates the flight time of the ion from the source to the detector with the m/z of the ion. c) Detector: The ion detector determines the mass of ions that are resolved by the mass analyzer and generates data which is then analyzed. The electron multiplier is the most commonly used detection technique. 5 1 2 3 4 5 Definitions of the components: Part 3 – MS-based detection techniques for PTMs 2. LC-MS/MS approach: LC-MS/MS a common analytical tool that combines physical separation by liquid chomatography with mass analysis and resolution by mass spectrometry. It is capable of separating and identifying complex mixtures for proteomics studies. a) Liquid chromatography: This is a chromatographic separation technique that separates molecules based on their differential adsorption and desorption between the stationary matrix phase in the column and the mobile phase. b) Affinity columns: Columns that make use of specific affinity interactions between the analyte of interest and the bound stationary phase matrix thereby successfully separating this component from a complex mixture. Immobilized Metal ion Affinity Chromatography (IMAC) is one such affinity technique that relies on the formation of specific coordinate-covalent bonds between certain amino acid residues of the protein (like histidine) and the immobilized metal ions. Phosphorylated proteins have been found to bind specifically to ions such as iron, gallium and zinc, thus facilitating their separation by IMAC. Recently, titanium dioxide (TiO2) columns have proved to be extremely useful for specific separation of phosphorylated proteins. c) Tandem MS: This is a mass spectrometry technique that makes use of a combination of ion source and two mass analyzers, separated by a collision cell, in order to provide improved resolution of the fragment ions. The mass analyzers may either be the same or different. The first mass analyzer usually operates in a scanning mode in order to select only a particular ion which is further fragmented and resolved in the second analyzer. This can be used for protein sequencing studies. 1 Part 2, Step 3 (a) MALDI-TOF analysis – Digestion & sample spotting Protein Trypsin digestion Matrix 2 PTM modified protein of interest Digested protein 3 4 196 –well MALDI Plate Action Description of the action 5 As shown in animatio n. PLEASE REDRAW ALL IMAGES. First show the tube on the left followed by appearance of the arrow and then the tube on the right. The hand must be shown to enter this tube and must place a drop of liquid from this tube onto the grey plate shown below. Once this happens, that spot must be zoomed into to show the image above. Audio Narration Post translational modifications can be detected by means of mass spectrometry due to the unique fragmentation patterns of phosphorylated seine and threonine residues.. The modified protein of interest is digested into smaller peptide fragments using a suitable enzyme like trypsin. This digest is then mixed with a suitable organic matrix such as a-cyano-4hydroxycinnamic acid, sinapinic acid etc. and then spotted on to a MALDI plate. 1 Part 2, Step 3 (b) MALDI-TOF analysis – Ionization & detection Laser Matrix & analyte 2 Detector Flight tube 3 Target plate MALDI 4 Action Description of the action 5 As shown in animatio n. Audio Narration The target plate containing the spotted matrix and analyte is placed First show the entire grey apparatus with all the in a vacuum chamber with high voltage and short laser pulses are labels and the violet rectangle laser source. Next applied. The laser energy gets absorbed by the matrix and is show a beam emerging from the rectangular box transferred to the analyte molecules which undergo rapid and falling on the white semicircular region. Once this happens, the colored ions must emerge from the sublimation resulting in gas phase ions. These ions are accelerated and travel through the flight tube at different rates. The lighter ions white surface as shown in animation. Once the move rapidly and reach the detector first while the heavier ions colored circles have appeared, they must move migrate slowly. The ions are resolved and detected on the basis of towards the ‘detector’. The smallest blue circles must move the fastest followed by the orange circles their m/z ratios and a mass spectrum is generated. and then the green circles. 72 32 81 107 25 72 80 Da 53 65 Observed peptide masses 80 Da implies 152 of 1 presence 192 81phosphate group! 107 152 164 25192 53 143 92 122 Presence of 2 151 170 65 164 phosphate group! 143 92 122 151 170 m/z 160 Da 81 107 32 143 92 122 151 170 25 m/z 3 Superimposed image 164 53 65 Relative abundance Expected peptide masses Relative abundance 2 MALDI-TOF analysis – Data interpretation Relative abundance 1 Part 2, Step 3 (c) m/z Thus, examination and comparison of list of observed peptide masses with expected peptide masses enables simple detection of PTMs. 4 Action Description of the action 5 As shown in animatio n. First show the graphical representation on the left followed by the graph on the right. These two must then merge together and be superimposed to give the third figure as shown in animation. The pink text and double headed arrows must then appear followed by the blue arrows and pink text boxes shown on the right. Finally the text below must appear. Audio Narration Identification of PTMs by MS largely lies in the interpretation of results. Comparison of the list of observed peptide masses from the spectrum generated with the expected peptide masses enables identification of those peptide fragments that contain any PTM due to the added mass of a modifying group. In this hypothetical example, two peptide fragments are found to have different m/z values, differing by 80 daltons and 160 daltons. It is known that the added mass of a phosphate group causes an increase in m/z of 80 daltons. Therefore, this principle of mass difference enables detection of modified fragments. 1 Part 2, Step 4 (a) LC-MS/MS based approach – Liquid chromatography Buffer Buffer solution 2 solution 2 3 PO43- Phosphorylated protein remains bound Metal ions Miniaturized immobilized metal affinity columns Direction of migration PO43- M3+ M3+ PO43M3+ IMAC: Ga3+, Zn2+, Fe3+ Other affinity columns: TiO2 Sample protein mixture Purified phosphorylated protein 4 Action Description of the action 5 Phosphorylated residue of protein As shown in animatio n. First show the ‘protein mixture’ along with the ‘column’. This mixture should be poured into the column after which the ‘buffer solution’ must be placed on the column. Liquid must flow into the column in the direction indicated. The column must be zoomed into to show the inset image on the right where the green and violet circles must move towards each other, along with the text below. Next, the pink and brown shapes must move from the column into the tube below after which the ‘buffer solution’ must be replaced (change color). Finally the green figure must also move down from the column into the tube below. TiO2 columns were found to have better selectivity and sensitivity of detection for phosphorylated peptide binding when compared to IMAC. Audio Narration Liquid chromatography coupled with mass spectrometry serves as a useful technique for enrichment and identification of proteins having a particular type of PTM from a complex mixture. The complex protein sample is loaded onto a miniaturized affinity column which will interact specifically with proteins having the PTM of interest. Here, we depict the use of immobilized metal affinity chromatography columns containing ions such as Ga3+, Zn2+, Fe3+ or TiO2 which have been found to specifically chelate the phosphorylated proteins. Unwanted proteins are removed by washing the column with a suitable buffer solution after which the phosphorylated protein of interest is eluted out by modifying the buffer solution. 1 2 Part 2, Step 4 (b) LC-MS/MS based approach – Tandem mass spectroscopy Purified protein Digested protein MS/MS analysis Analysis and interpretation of data is carried out as described for MALDITOF-MS. LASER Detector 3 TOF 2 (RF mode) TOF 1 (scanning mode) 4 Collision cell Action Description of the action 5 Reflector As shown in animatio n. Audio Narration First show the reaction on top appearing after which an arrow from The protein purified by liquid chromatography is then the tube on the right must indicate that this sample enters the subjected to typtic digestion followed by analysis using tandem instrument below. Show the instrument below as depicted followed mass spectrometry. Here we demonstrate the use of MALDIby appearance of a red beam from the red box which must strike TOF-TOF-MS for resolution of the generated ion fragments. the dotted line. Next, the colored circles must appear which must Separation is based on the flight time of the ions and greater move into the pink box with the smallest moving the fastest & vice resolution is achieved due to the presence of two mass versa. The yellow circle must then move to the other end of the analyzers. The peptide ion spectrum generated is analyzed by ‘collision cell’ followed by appearance of three smaller circles and comparing it with the expected spectrum, thereby allowing disappearance of the large yellow circle. These smaller circles must determination of modified peptides having different m/z values. move towards the detector, with the smallest moving the fastest. Master Layout (Part 4) 1 2 This animation consists of 4 parts: Part 1: Overview of PTMs Part 2: Gel-based detection techniques for PTMs Part 3: MS-based detection techniques for PTMs Part 4: Microarray-based detection techniques for PTMs Autoradiography film [g-33P] ATP ADP Protein array Kinase enzyme 3 Phosphorylated proteins 1. Protein microarrays Array scanning ` 4 Antibody microarrays Phosphorylated proteins 5 Labeled protein mixture 2. Antibody microarrays 1 2 Definitions of the components: Part 4 – Microarray-based detection techniques for PTMs 1. Protein microarrays: These are miniaturized arrays normally made of glass, onto which small quantities of many proteins can be simultaneously immobilized and analyzed. For detection of phosphorylation sites, potential protein substrates are immobilized on to the array. a) Kinase enzyme: An enzyme that is responsible for phosphorylation of specific amino acid residues in the protein with the help of ATP as a phosphate donor. 3 4 5 b) Phosphorylated proteins: Proteins that have been phosphorylated at specific amino acid residues. c) Autoradiography: Radioactivity is the process by which certain elements spontaneously emit energy in the form of particles or waves due to disintegration of the unstable atomic nuclei into a more stable form. These radiations that are given out can be detected by means of autoradiography, wherein the radiations are allowed to strike a photographic film which on exposure shows the presence radioactive emissions. 1 2 3 4 5 Definitions of the components: Part 4 – Microarray-based detection techniques for PTMs 2. Antibody microarrays: An array onto which different antibodies are spotted, which have specific binding domains for detection of the protein of interest from a complex mixture. For detection of PTMs, antibodies against specific protein motifs containing the PTM or against a specific residue containing a phosphorylated site may be used. a) Labeled protein mixture: The protein mixture containing the protein of interest is labeled uniformly with a suitable fluorescent dye which can be detected by scanning at the appropriate wavelength. Cyanine dyes are commonly used for such labeling purposes. b) Array scanning: Once the binding interactions have taken place on the array surface and excess unbound material has been washed away, the array is scanned using a microarray scanner. This scans the array at a suitable wavelength depending upon the fluorescent dye used for labeling purposes to generate an image depicting the array positions at which binding has occurred. 1 Part 4, Step 1 (a) Protein microarrays 2 Ser [g-33P] ATP ADP Ser Kinase enzyme 3 Protein substrate Proteome array containing potential substrates for phosphorylation Phosphorylated protein 33P] ATP [g-Kinase enzyme solution 4 Action Description of the action 5 As shown in animatio n. First show the ‘proteome array surface’ along with the flast with pink solution on the right. The hand must appear and pipette out the solution from the flask on to the array surface such that the entire surface gets covered with a layer of pink solution. Next, the blue solution must appear and similar procedure must be carried out with that. Once this is done, one of the spots on the array surface must be zoomed into to depict the inset image on the right hand side in which the reaction components must appear sequentially as shown in animation. Audio Narration PTMs can also be detected by means of protein microarrays using a kinase assay. Potential substrates for protein phosphorylation are immobilized on a suitably coated array surface. To this, kinase enzyme and gamma P-32 labeled ATP are then added and the array is incubated at 30oC. The phosphorylation reaction occurs at those sites containing proteins that can be modified. 1 Part 4, Step 1 (b) Phosphorylated proteins Protein microarrays Washing 2 Proteome array P 33 P 33 P 33 P 33 3 P 33 Radioactive emissions 4 Developed image Action Description of the action 5 As shown in animatio n. First show the label ‘washing’ and disappearance of the blue and pink solutions coating the array surface and only certain sites as shown must contain a star label. Next a gray surface must appear and the black surface must be placed in contact with this gray surface as shown. This must be zoomed into to show the inset on the left along with the animation as depicted. DetectionAutoradiography film Audio Narration After sufficient incubation, excess unbound ATP and enzyme are washed off the array surface. Detection is carried out by means of autoradiography wherein a photographic film is placed in contact with the array surface. The radioactive emissions from the phosphate label present at the phosphorylated protein sites strike the film. Upon development, the positions at which phosphorylation has occurred can be clearly determined. Thus proteome chip technology offers a useful platform for detection of phosphporylated proteins. 1 Part 4, Step 2 (a) Antibody microarrays Labeled protein mixture added 2 3 Polymer coated glass slide Anti-phospho serine, threonine and tyrosine antibodies immobilized on array Specific binding of phosphorylated proteins 4 Action Description of the action 5 As shown in animatio n. First show the gray surface (which must be horizontal) on the left followed by binding of the ‘Y’ shaped brown objects as shown. Next, the orange cloud along with the flagged shapes must appear on top of this surface. The green circles must be shown to bind to the brown shapes as depicted while the rest must remain unbound. Audio Narration Antibodies specific to phosphorylated serine, threonine or tyrosine residues as well as motif antibodies can be immobilized on to a suitably coated microarray surface and used for detection of PTM. The complex protein mixture containing modified and unmodified proteins is labeled with a suitable fluorescent tag molecule and added to the array surface. Specific binding interactions occur between the phosphorylated proteins and their corresponding antibodies. 1 Part 4, Step 2 (b) Antibody microarrays Microarray scanner Array washing Unbound proteins removed 2 3 Bound phosphorylated proteins Array image 4 Action Description of the action 5 As shown in animatio n. Audio Narration Please re-draw all images. The blue solution must appear on The array is then washed to remove any excess top of the existing components and must then be removed unbound proteins from the surface. This is followed by from the array surface such that only the green circles remain scanning of the array using a microarray scanner at a bound to the surface as shown. The ‘microarray scanner’ suitable wavelength to detect the fluorescent tag of the image must then appear and the array must shrink and be bound proteins. This method offers sensitive and placed on the extended grey region as depicted in animation. simultaneous detection of large number of post Next the orange beam must appear which must fall on the translationally modified proteins. array surface followed by appearance of the computer with the image pattern as shown. 1 Interactivity option 1:Step No: 1 Based on the two MALDI-TOF-MS spectrums shown below, determine the total number of phosphorylated sites present in the protein. 3 77 40 25 81 107 164 53 96 122143 151 170 62 Relative abundance 2 Relative abundance Expected peptide masses Observed peptide masses 157 25 4 A) 4 Interacativity Type 5 Choose the correct answer. 133 62 m/z Answers: 200 81 164 96 122143 170 151 m/z B) 6 Options C) 5 D) 7 Results The correct answer is B. Once the user User must be shown the two graphs on top followed by the chooses an answer, a sign must first be question below and the answers.displayed saying ‘correct’ or ‘incorrect’ User must be allowed to choose depending on the answer given. Next, the colored boxes along with the double one of the four options. headed arrows must appear sequentially as depicted in animation. 267 1 Questionnaire 1. Proline most commonly undergoes which of the following PTMs? Answers: a) Glycosylation b) Phosphorylation c) Hydroxylation d) Acylation 2 2. N-linked glycosylation most commonly occurs on which of the following amino acids? Answers: a) Serine b) Threonine c) Aspartic acid d) Aspargine 3. Which of the following fluorescent dyes is used for specific detection of phosphoproteins? 3 Answers: a) Pro-Q-diamond b) SYPRO-Ruby Red c) SYPRO-Ruby Orange d) Coomassie brilliant blue 4 4. Difference in molecular weight of 240 Da between two peptide fragments obtained by MALDI-TOF-MS is indicative of presence of how many phosphorylation sites on the protein? Answers: a) 2 b) 3 c) 0 d) 5 5 5. Which of the following metal ions is not used for separation of phosphorylated proteins during liquid chromatography? Answers: a) Zn2+ b) Fe3+ c) Ga3+ d) K+ Links for further reading Research papers: • • • • • • • • • • Berggren, K. N. et al. An improved formulation of SYPRO Ruby protein gel stain: Comparison with the original formulation and with a ruthenium II tris (bathophenanthroline disulfonate) formulation. Proteomics 2002, 2, 486-498. Reinders, J. & Sickmann, A. Modificomics: Posttranslational modifications beyond protein phosphorylation and glycosylation. Biomolecular Engineering 2007, 24:169-177. Zhang, H. et al., Phosphoprotein Analysis Using Antibodies Broadly Reactive against Phosphorylated Motifs. J Biol. Chem. 2002, 277(42):39379-39387. Delom, F. & Chevet, E. Phosphoprotein analysis: from proteins to proteomes. Proteome Science 2006, 4:!5. Andersson L, Porath J: Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem1986, 154:250-254. Posewitz MC, Tempst P: Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem 1999, 71:2883-2892. Neville DC, Rozanas CR, Price EM, Gruis DB, Verkman AS, Townsend RR: Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix assisted laser desorption mass spectrometry. Protein Sci 1997, 6:2436-2445. Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ: Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 2005, 4:873-886. Oda Y, Nagasu T, Chait BT: Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat Biotechnol 2001, 19:379-382. Janek K, Wenschuh H, Bienert M, Krause E: Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom 2001, 15:1593-1599. Links for further reading Research papers: • • • • • Wilm M, Mann M: Analytical properties of the nanoelectrospray ion source. Anal Chem 1996, 68:1-8. Erdjan Salih, Phosphoproteomics by mass spectrometry and classical protein chemistry approaches. Mass Spectrometry Reviews 2005, 24:828-846. Morelle, W. et al., The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics 2006, 6:3993-4015. Ptacek, J. et al., Global analysis of protein phosphorylation in yeast. Nature Letters 2005, 438:679-684. Zhu, H. et al., Global Analysis of Protein Activities Using Proteome Chips. Science 2001, 293:2101-2105. Books: • Biochemistry by A.L.Lehninger et al., 4th edition • Post-Translational Modification: Phosphorylation and Phosphatases. Current Protocols in Protein Science Supplement 31, Chapter 13. (Cold Spring Harbour Proteomics Manual) • Post-Translational Modification: Glycosylation. Current Protocols in Protein Science Supplement 26, Chapter 12. (Cold Spring Harbour Proteomics Manual) Websites: • Steinberg et al. Pro-Q Diamond phosphoprotein stain: a new reagent for detection of phosphoproteins and phosphopeptides in polyacrylamide gels and in microarrays. • http://www.biomax.us/antibody-arrays.php