* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Part 2 Notes and Notes Questions
Survey
Document related concepts
Transcript
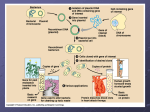
Name: ____________________________________________ Date: ___________________________ Period: _____ Unit 8 Notes, Part 2: Biotechnology AP Biology, Mrs. Krouse A. How DNA from two organisms be compared? 1. With our current technology, scientists can determine the sequence of nitrogen bases in DNA segments that differ greatly from organism to organism. 2. Another way to compare the DNA from two individuals is by using gel electrophoresis. Gel electrophoresis can be used to produce a unique DNA fingerprint for each person. 3. Before the DNA fingerprint can be created however, the DNA double helix must be “cut” to produce fragments of DNA that will form the “bands” of the fingerprint. 4. The DNA molecule is cut using enzymes taken from bacteria called restriction enzymes (aka restriction endonucleases). In bacteria, the function of these enzymes is to protect the bacterium by cutting up foreign DNA from other organisms or viruses (aka phages). 5. Each specific restriction enzyme recognizes a particular DNA sequence (aka restriction site) and cuts both DNA strands at particular places within the sequence. For example, the restriction site for the restriction enzyme EcoR1 is given below with the cut location shown in green: Note: Most restriction sites are symmetrical / pallindromic (i.e. the sequence of nucleotides is the same on both DNA strands when they are read in the 5’ to 3’ direction). Because restriction sites are typically short (usually four to eight nucleotides in length), they occur by chance many times in a long DNA molecule, so a particular restriction enzyme will make a large number of cuts along the length of the DNA molecule. This will produce many smaller “chunks” of DNA called restriction fragments. A particular DNA molecule cut by the same restriction enzyme will always produce the same set of restriction fragments. A change in the DNA sequence at a restriction site is called a restriction fragment length polymorphism (RFLP) and may prevent a restriction enzyme from cutting at this site, resulting in restriction fragments of different lengths. 6. After two DNA molecules from different organisms have been cut using the same restriction enzyme, they are “loaded” into two small divets called wells at one end of a thin slice of agarose gel. 7. Once the DNA samples are loaded, an electrical current is run through the gel. The well end has an overall negative charge, and the opposite end has an overall positive charge. 8. Because DNA molecules have an overall negative charge (due to negatively-charged phosphate groups), DNA loaded into the wells will migrate through the gel to the positive end (because opposite charges attract) 9. Because the gel is a thick mesh, smaller / shorter fragments migrate faster than long ones. After the current is turned off, the different fragments can be viewed as “bands,” with larger fragments closer to the well-end and smaller fragments closer to the far end. To see the bands, a DNA-binding dye must be added that fluoresces under UV light. 10. If the two DNA molecules loaded into the two wells were initially cut with the same restriction enzyme, then the different band patterns indicate that they came from different sources. B. How can we actually see DNA bands if DNA molecules are very small? 11. The DNA samples must be copied (aka amplified) many times in order to be viewed as a thick band on a gel. 12. If scientists cut a DNA sample with a restriction enzyme, this will produce “overhangs” of single stranded DNA at the cut site. These “overhangs” are called “sticky ends.” If a bacterial plasmid (a small circle of DNA outside the bacterial chromosome) is cut with the same restriction enzyme, it will have complementary “sticky” ends with the DNA sample and they can be joined using the enzyme DNA ligase. 13. This new plasmid (with DNA from another sample) is considered recombinant DNA because it contains DNA from two different organisms. 14. If we are able to induce a bacterial cell to take in the newly-created recombinant plasmid (technique described in #21), the bacteria can copy (aka amplify) a gene for us). Every time the bacterial cell replicates its plasmid DNA before reproduction by binary fission (cell splitting from one parent cell to two daughter cells), it also replicates the DNA sample of interest. This is called “gene cloning” and it can produce many copies of a particular DNA sequence to be loaded into a well for gel electrophoresis. 15. DNA sequences can be copied without the use of bacteria using polymerase chain reaction (PCR). This technique can make billions of copies of a target segment of DNA in a few hours in a test tube. 16. In the first step of PCR, the DNA is heated to denature (separate) the DNA strands and then cooled. Once the DNA mixture is cooled, single-stranded DNA primers that are complementary to sequences on opposite strands at the ends of the target sequence anneal (hydrogen bond) to these target sequences. 17. A heat-stable form of DNA polymerase (Taq polymerase) extends the primers to copy the sequences in the 5’ to 3’ direction. A heat-resistant form of DNA polymerase must be used so that it does not denature during the first step of PCR. Taq polymerase is taken from a bacterial species, Thermus aquaticus, that lives in hot springs, so it must have a heat-resistant DNA polymerase for its own DNA replication. 18. This process can be repeated to exponentially increase the number of copies of a particular DNA sequence. C. How can we create bacteria that have been “transformed” by recombinant DNA? 19. Remember, bacterial transformation involves a bacterium taking in “naked DNA” from its environment. 20. Let’s say we want to insert a gene from an organism (ex: the human insulin gene) into a bacterial plasmid using the method described in #12. The product will be a recombinant plasmid, meaning it contains DNA from two different organisms (a human and a bacterium). The process of creating the recombinant plasmid happens outside the cells of both organisms, so the recombinant plasmid is considered “naked DNA.” 21. We can “transform” a bacterial cell by forcing it to “take up” the naked recombinant plasmid we have created. We do this by placing the bacterial cell in a calcium chloride (CaCl 2) solution and “shocking” it with a lot of heat. The combination of these two factors causes holes to form in the bacterial cell’s membrane, through which the plasmid DNA can enter the bacterial cell. 22. When the “transformed” bacterium replicates its plasmid DNA before reproduction by binary fission (cell splitting from one parent cell to two daughter cells), it also replicates the human insulin gene. This is called “gene cloning” and it can produce many copies of a particular gene. 23. When the “transformed” bacterium transcribes and translates its own plasmid DNA, it also transcribes and translates the human insulin gene to produce human insulin protein. This human insulin protein can be “harvested” and used to treat diabetes. 24. Below is a summary diagram showing the process of bacterial transformation and some uses/applications of this technology. D. How might the AP test assess my knowledge of bacterial transformation? Two sample scenarios and sets of practice question are given below: A scientist is using an ampicillin-sensitive strain of bacteria that cannot use lactose because it has a nonfunctional gene in the lac operon. She has two plasmids. One contains a functional copy of the affected gene of the lac operon, and the other contains the gene for ampicillin resistance. Using restriction enzymes and DNA ligase, she forms a recombinant plasmid containing both genes. She then adds a high concentration of the plasmid to a tube of the bacteria in a medium for bacterial growth that contains glucose as the only energy source. This tube (+) and a control tube (-) with similar bacteria but no plasmid are both incubated under the appropriate conditions for growth and plasmid uptake. The scientist then spreads a sample of each bacterial culture (+ and -) on each of the three types of plates indicated to the right. 25. If no new mutations occur, it would be most reasonable to expect bacterial growth on which of the following plates? (A) 1 and 2 only (B) 3 and 4 only (C) 5 and 6 only (D) 4, 5, and 6 only (E) 1, 2, 3, and 4 only 26. If the scientist had forgotten to use DNA ligase during the preparation of the recombinant plasmid, bacterial growth would most likely have occurred on which of the following? (A) 1 and 2 only (B) 1 and 4 only (C) 4 and 5 only (D) 1, 2, and 3 only (E) 4, 5, and 6 only In a transformation experiment, a sample of E. coli bacteria was mixed with a plasmid containing the gene for resistance to the antibiotic ampicillin (ampr). Plasmid was not added to a second sample. Samples were plated on nutrient agar plates, some of which were supplemented with the antibiotic ampicillin. The results of E. coli growth are summarized below. The shaded area represents extensive growth of bacteria; dots represent individual colonies of bacteria. Note: The shaded areas (Plates I and III) show extensive growth of bacteria and are called “bacterial lawns.” The area with “dots” (Plate IV) shows individual colonies of bacteria. The bacteria in these colonies have been successfully “transformed” and are able to live in an environment with ampicillin. Not all bacteria will be successfully transformed, and untransformed bacteria will die, leaving blank spaces surrounding the colonies on the plate. 27. Plates that have only ampicillin-resistant bacteria growing include which of the following? (A) I only (B) III only (C) IV only (D) I and II 28. (A) (B) (C) (D) Which of the following best explains why there is no growth on plate II? The initial E. coli culture was not ampicillin-resistant. The transformation procedure killed the bacteria. Nutrient agar inhibits E. coli growth. The bacteria on the plate were transformed. 29. (A) (B) (C) (D) Plates I and III were included in the experimental design in order to demonstrate that the E. coli cultures were viable demonstrate that the plasmid can lose its ampr gene demonstrate that the plasmid is needed for E. coli growth prepare the E. coli for transformation E. How can recombinant DNA technology and bacterial transformation be used commercially or medically? 30. As mentioned before, we can cause bacteria to produce the human insulin protein by creating recombinant DNA plasmids containing the human insulin gene. We can use the same method with other human proteins like human growth hormone. 31. Scientists can also use bacteria as “vectors” to introduce foreign genes into other organisms (ex: plants and animals). For example, Agrobacterium tumefaciens is a species of bacteria that infects plant cells and causes the formation of tumors. This condition is called crown gall disease. During infection, a section of a plasmid found within the bacterium (called the Ti plasmid) integrates itself into the plant cell genome. Therefore, scientists can insert a gene from any organism into the Ti plasmid and use the plasmid to deliver the gene to the plant cells. An example of this is golden rice, which has been modified with the gene for beta carotene (Vitamin A) synthesis. This is an extremely beneficial crop in areas of the world where people have vitamin A deficiencies and good conditions for growing rice but not other beta carotene-rich foods (ex: carrots and sweet potatoes). Notes Questions 1. What is the purpose of polymerase chain reaction (PCR)? 2. What are the advantages of using PCR vs. gene cloning inside a bacterial plasmid? 3. What are restriction fragment length polymorphisms (RFLPs) and how can they be studied using gel electrophoresis? 4. Which of the following sequences in double-stranded DNA is most likely to be recognized as a cutting site for a restriction enzyme? Explain your answer. A) A A C G T T G C B) C C C C G G G G C) A G G A T C C T D) A T A T T A T A 5. A restriction fragment analysis was carried out on DNA samples taken from members of a family due to questionable paternity of one of the family’s children. The results of the gel generated for analysis are shown to the right. Which of the following statements is supported by the data? Explain your answer. (A) Both children are related to both parents. (B) Child I is related to the man but child II is not. (C) Both children are unrelated to either of the parents. (D) Child II is related to the man but child I is not. 6. For the DNA segment that is cut at restriction sites I and II shown on the image below, draw where fragments A, B, and C will be located on a gel after electrophoresis. How do you know where each fragment will “end up”? Gel: Negative End (Well End) Positive End 7. List the correct order for the following steps used in the creation of recombinant plasmids and bacterial transformation. I. II. III. IV. V. Transform bacteria with a recombinant DNA molecule. Cut the plasmid DNA using restriction enzymes. Extract plasmid DNA from bacterial cells. Hydrogen-bond the plasmid DNA to nonplasmid DNA fragments. Use ligase to seal plasmid DNA to nonplasmid DNA. In a molecular biology laboratory, a student obtained E. coli bacteria cells and used a common transformation procedure to induce the uptake of plasmid DNA with a gene for resistance to the antibiotic kanamycin. The results below were obtained. 8. What results do we see on Plate I and why? 9. What results do we see on Plate II and why? 10. What results do we see on Plate III and why? 11. What results do we see on Plate IV and why?