* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Sequence-specific assignments
Survey
Document related concepts
Transcript
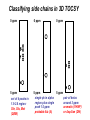
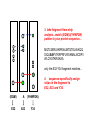
Resonance assignments Part II: Approaches to sequence-specific assignments Sequence-specific assignments • • • • • suppose we have the sequence of our protein from some independent measurement suppose we’ve assigned an isoleucine spin system, and there’s only one isoleucine in the sequence (unique), at position 48. Then we know our isoleucine is Ile48. there won’t be very many unique amino acid residues in a protein, however. but there will be many unique dipeptide sequences (or tripeptide etc...) but in order to use this fact, we need to be able to connect adjacent residues. unique residues (arrows) and unique dipeptide sequences in lac repressor Linking spin systems using nOe’s • because the nOe depends upon interatomic distance and not upon J coupling, it can be used to connect spin systems which are adjacent in space but not part of the same spin system, for instance two residues adjacent in the sequence •general nomenclature for interatomic distance between atoms A and B in residues i and j: dAB(i,j) • nOe correlations are denoted using the distance nomenclature, e.g. “dbN(i,i+1) nOe” or “dbN (i,i+1) correlation” • daN(i,i+1), dNN(i,i+1), and sometimes dbN(i,i+1) are used to connect adjacent residues The 2D NOESY pulse sequence from Glasel & Deutscher p. 354 mixing period tm between t1 and t2 allows for cross-relaxation between nuclei (mostly zero quantum as we’ve seen) --> result is crosspeaks due to nOe 2D NOESY: linking spin systems 4.HN/5.HN 5.HN/6.HN diagonal: no magnetization transferred crosspeaks: intersection of chemical shifts of atoms which are close in space, i.e. < 5 Å 1H 6.HN/7.HN 3.HN/4.HN 1H amide-amide region of 2D NOESY of P22 Cro protein, showing dNN(i,i+1) correlations-can “walk” along the chain from one residue to the next. Residues 3-7 shown. Classic 1H resonance assignment protocols • Sequential assignment method (Wuthrich) A method in which one first makes the spin-system assignments, followed by sequence-specific assignment using unique fragments of sequence. • Main-chain directed assignment method (Englander). This alternative technique does not focus on assigning all the spin systems first. Rather, it focuses on the backbone and links sizable stretches of backbone residues via sequential (i,i+1) nOe’s and other nOe’s that are characteristic of secondary structures (more on this in a second). This technique is particularly useful when there is some knowledge of secondary structure beforehand. Summary of sequential approach 1. assign most or all spin systems Arg Tyr Ser Ala Ala Asn Trp 3. assemble larger sections of sequence-specific assignments from dipeptide fragments, until the whole protein has been assigned 2. connect adjacent spin systems using backbone nOe’s to identify unique dipeptides “backbone” refers to alpha and amide protons Summary of main-chain directed approach 1. assign a few unique spin systems and use as entries onto the backbone Arg Tyr Ser Ala Ala Asn Trp 3. fill in missing spin system assignments 2. walk down the backbone using sequential and other backbone nOe’s “backbone” refers to alpha and amide protons Close interatomic distances in secondary structures parallel beta-sheet antiparallel beta-sheet alpha-helix type I turn type II turn Close interatomic distances in 2ndary structures nOes and secondary structures residue # • In NMR papers you’ll sometimes see charts like the one shown above. A thick bar means a strong nOe (short distance), a thin bar means a weak nOe (long but still visible distance) • The fact that certain nOe’s are characteristic of secondary structures allows one to make secondary structure assignments more or less concurrently with sequential assignments. As we will soon see, coupling constants and chemical shifts also aid in secondary structure assignment ...you can see that it would be easiest to link adjacent residues in helices with sequential amide-amide nOe’s, whereas in beta sheets (strand) sequential alpha-amide nOe’s are stronger d~2.8 Å d~2.2 Å Modern assignment methods that use heteronuclear shift correlation • • • for larger proteins (>10-15 kD), assignment methods based on the 2D homonuclear 1H-1H correlation methods (COSY/TOCSY/NOESY) that we’ve been discussing don’t work very well because of overlapping resonances and broad linewidths. an alternative (which is now used even for small proteins in most cases) is to use heteronuclear shift correlation experiments on 13C, 15N labelled samples. in these experiments, magnetization is transferred between 1H, 13C and /or 15N through large one-bond or in some cases two-bond scalar couplings. Scalar couplings commonly used in heteronuclear shift correlation all couplings are in units of Hz 15N-1H HSQC based techniques •as we have seen, one of the simplest types of heteronuclear shift correlation is the HSQC experiment, which correlates 1H chemical shift to the chemical shift of a 15N or 13C connected by a single bond •2D heteronuclear shift correlation can be combined with homonuclear experiments such as 1H-1H 2D NOESY or 2D TOCSY to yield 3-dimensional spectra 3D HSQC-TOCSY CO2CH2 H CH2 N C C H O H CH3 N C C H O 2 of the dimensions are HN correlation (HSQC) 3rd dimension is 1H-1H TOCSY correlations from the HN proton 3D HSQC-NOESY CO2CH2 H CH2 N C C H O H CH3 N C C H O Like 3D TOCSY but includes interresidue and interspin system correlations (dashed lines). 3D HSQC-NOESY and HSQC-TOCSY these planes can be thought of as a 15N-1H HSQC the planes (parallel to the slide) can be thought of as a 1H-1H NOESY (1H) NOESY dimension the 15N shift dimension can resolve peaks that would overlap in a 2D NOESY 15N dimension HN 1H view of a 3D NOESY experiment dimension Analyzing 3D spectra rather than try to look at this whole thing at once NOESY or TOCSY (1H) dimension 15N dimension HN 1H dimension look at vectors or “strips” corresponding to peaks on an HSQC (particular 15N and HN shift combinations)--> NOESY/TOCSY correlations will be along the length of the strip Extracting strips in a 3D Use 2D HSQC as reference spectrum Strip of 3D corresponding to peak in HSQC 0 ppm F2:120 ppm, F3: 8.0 ppm 15N (F2 in 3D) F2 = 120 ppm (plane of paper) crosspeaks to side chain 1H F1: 8.1-7.9 crosspeak to alpha NOESY or TOCSY dimension diagonal peak (amide region) HN (F3 in 3D) want to look at TOCSY or NOESY correlations from the amide proton corresponding to this HSQC peak 10 ppm 8.1-7.9 ppm HN dimension (F3) Classifying side chains in 3D TOCSY 0 ppm 0 ppm 0 ppm b b g b a a a 5 ppm set of 4 peaks in 1.9-2.6 region: Gln, Glu, Met (QEM) 5 ppm 5 ppm single pk in alpha pair of betas region plus single around 3 ppm: peak 1-2 ppm: aromatic (YHWF) probable Ala (A) or Asp/Asn (DN) Using 3D TOCSY/NOESY dual strip analysis 3D TOCSY 3D NOESY b daN(i,i+1) b dbN(i,i+1) g same residue b a different residue a (EQM) A (YHWHDN) TOCSY --> intraresidue xpks 1. spin system classifications (EQM) A (YHWHDN) NOESY --> interresidue xpks--> 2. connect strips into sequence fragments 3. take fragment from strip analysis...match (EQM)A(YHWFDN) pattern to your protein sequence... MQTLSERLKKRRIALMTQTELAVKQQ SIQLIEAYVTKRPRFLFEIAMALNCDPV WLQYGTKRGKAA only the E32-Y34 fragment matches... 4. sequence specifically assign strips in the fragment to E32, A33 and Y34. (EQM) A (YHWFDN) E32 A33 Y34 5. annotate the corresponding 2D HSQC peaks with the new assignments E32 A33 15N (F2 in 3D) Y34 HN (F3 in 3D) 6. proceed until entire HSQC is assigned... Triple-resonance experiments • • • • there is a whole raft of experiments that use both 13C and 15N correlations to 1H nuclei the beauty of these experiments is that they can connect adjacent residues without requiring any nOe information-it’s all through-bond scalar coupling interactions. Makes sequence-specific assignment more reliable. they also use mostly one-bond couplings, which aren’t very sensitive to the protein conformation (unlike, say, three-bond couplings, which vary significantly with conformation, as we will see) limiting factors: 13C is expensive and these exp’ts can be tricky Beyond “spin systems”: connecting residues using heteronuclear J couplings -7 Hz H H N C C R O 11 Hz H H N C C R O H H N C C R O the HNCA experiment above connects the HN group to the alpha carbon of both the same residue and the previous one. The two-bond N-C coupling traverses the carbonyl group, which is a barrier to using 1H-1H scalar couplings to connect residues