* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Hybrid
Survey
Document related concepts
Transcript
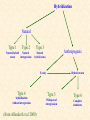
Lecture VI Molecular Ecology (Hybridization) What is Hybridization? Hybrid – Oxford Dictionary of Zoology – - an individual animal that results from a cross between parents of differing genotypes. Strictly, most individuals in an outbreeding population are hybrids, but the term is more usually reserved for cases in which the parents are individuals whose genomes are sufficiently distinct for them to be recognized as different species or subspecies. A good example is a mule, produced by cross-breeding an ass and a horse (each of which can breed as true species). Hybrids may be fertile or sterile depending on qualitative and/or quantitative differences in the genomes of the two parents. Hybrids, like the mule, whose parents are of different species, are frequently sterile. How important is hybridization? Why devote a whole lecture to it? The theoretical and empirical study of hybridization is somewhat inseparable from that of speciation. Remember the BSC (Biological Species Concept). If reproductive islolation should define a species, then what does the existence of hybridization mean? Does it mean that two lineages that hybridize are not species? Hybridization is one of the primary problems in applying the BSC, the other being that knowledge on reproductive isolation (or, inability to hybridize) is often not available. For fishes, this issue is even more prominent as not only do many fishes hybridize naturally in nature, but those that don‘t will often do so under artificial conditions, and even more often if given a „little help“ in the laboratory. Hybrid – Oxford Dictionary of Zoology – - an individual animal that results from a cross between parents of differing genotypes. Strictly, most individuals in an outbreeding population are hybrids, but the term is more usually reserved for cases in which the parents are individuals whose genomes are sufficiently distinct for them to be recognized as different species or subspecies. A good example is a mule, produced by cross-breeding an ass and a horse (each of which can breed as true species). Hybrids may be fertile or sterile depending on qualitative and/or quantitative differences in the genomes of the two parents. Hybrids, like the mule, whose parents are of different species, are frequently sterile. In practice, evolutionary biologists do not draw any „systematic lines“, when discussing hybridization. As you have learned, it is difficult enough to define species, with a „subspecies being even more controversial. Thus, in practice, the taxonomic level of the units described to hybridize is very loosely interpreted, and with domesticated lines, for example, one may related simply to a single trait, or genetic locus. Introgression – (the distinction between hybridization & introgression is very important!!) The incorporation of the genes of one species into the gene pool of another species. If the ranges of two species overlap and fertile hybrids are produced, the hybrids tend to backcross with the more abundant species. This results in a population in which most individuals resemble the more abundant parents but also possess some of the characteristics of the other parent species. Hybrid vigour (heterosis) – (primarily relevant in domesticated strains, and is probably mostly do to a „release“ from the effects of inbreeding) The increased vigour of growth, survival, and fertility of hybrids, as compared with the two homozygotes. It usually results from crosses between two genetically different, highly inbred lines. It is always associated wtih increased heterozygosity. Hybrids swarm – A continuous series of hyrids that are morphologically distinct from one another, which results from the hybridization of two species followed by the crossing and backcrossing of subsequent generations. The hybrids are very variable owing to segregation of alleles at each locus. Hybrid dysgenesis– A complex of genetic abnormalities, which occurs in certain hybrids. The abnormalities may include sterility, enhanced rates of gene mutations, and chromosomal rearrangements. Hybrid dysgenesis occurs in the hybrid offspring of certain strains of Drosophila, in which it is thought to be due to mutations induced by transposon-like elements. How Common is Hybridization in Fishes? Schwartz (1972, 1981) compiled a total of 3,759 references dealing with the natural and artificial hybridization of fishes. Schwartz FJ 1981. World Literature to Fish Hybrids, With an Analysis by Family, Species, and Hybrid: Supplement 1. NOAA Technical Report NMFS SSFR-750, US Dept. of Commerce 507 pp. Most of these studies were published after Hubbs (1955) summarized his extensive investigations of natural hybridization among North American fishes. Hubbs CL (1955) Hybridization between fish species in nature. Systematic Zoology 4, 1-20. Natural hybridization is believed to be more common in fishes than in other groups of vertebrates. Several characteristics of fishes may account for this distinction: 1) external fertilization 2) weak ethological isolating mechanisms, 3) unequal abundance of the two parental species, 4) competition for limited spawning habitat, 5) susceptibility to secondary contact between recently evolved forms These characteristics are affected to varying degrees by local habitat. I would add artificial rearing, transport and planned or accidental releases to this list. Of these characteristics, which are promoting by environmental changes, natural or man-made? Natural and man-induced changes in environmental conditions are often cited as causes of hybridization in fishes. For example, hybridization is relatively common among temperate freshwater fishes in areas where geologic and climatic across the Pleistocene have drastically altered aquatic environments, but hybridization appears to be rare among marine and tropical fishes that inhabit more stable environments. Man caused habitat changes in North America have also been correlated with hybridization between both previously allopatric and naturally sympatric pairs of species (Hubbs et al 1953, Nelson 1966, 1973, Stevenson & Buchanon 1973). In addition, sympatric species that rarely or never hybridize in nature often hybridize freely within the confines of aquaria (Hubbs 1955). As Hubbs's (1955) extensive investigations led him to conclude " It is evident that the hybridization is conditioned by environmental factors"- Thus, it is important to understand the role of the environment in hybridization. The problems with hybrids: setting conservation guidelines Allendorf FW, Leary RF, Spruell P & Wenburg JK (2002) Trends in Ecology & Evolution 16(11) 613-622 „rates of hybridization and introgression are increasing dramatically worldwide because of translocations of organisms and habitat modifications by humans. Hybridization has contributed to the extinction of many species through direct and indirect means. However, recent studies have found that natural hybridization has played an important role in the evolution of many plant and animal taxa. Determining whether hybridization is natural or anthropogenic is crucial for conservation, but is often difficult to achieve. Any policy that deals with hybrids must be flexible and must recognize that nearly every situation involving hybridization is different enough that general rules are not likely to be effective“ Hybridization Natural Type 1 Type 2 Natural hybrid Natural taxon introgression Type 3 Anthropogenic Natural hybrid zones F1 only Type 4 hybridization without introgression (from Allendorf et al 2000) Hybrid swarm Type 5 Type 6 Widespread introgression Complete admixture Recognize that if F1 hybrids are produced but there is no backcrossing, and no F2 generation, then hybridization is irrelevant for both evolution, and systematics. Thus, „hybridization“ is not neccessarily a problem for the BSC, and F1 hybridization should not be considered as evidence of the lack of reproductive isolation, but rather simply the lack of a prezygotic barrier. Prezygotic isolation: (prevents hybridization from occurring, but, how stable are such mechanisms in a changing environment?) Mechanisms preventing mating geographic isolation (very common in freshwater fishes) temporal isolation (very common in freshwater fishes) different mate recognition systems (common within some groups, like cichlids) mechanical isolation (copulation unsuccessful, no transfer of gametes) Mechanisms preventing fertilization gamete mortality genetic incompatibility (often when numbers of chromosomes differ, or other chromosome level differences, there is also cyto-nuclear incompatibility) Postzygotic isolation: Mechanisms preventing development of interspecific hybrids (thus the key issue of postzygotic mechanisms is to prevent hybridization) zygote mortality (e.g. Dobzhansky-Muller incompatibilities) hybrid inviability / sterility METHODS OF DETECTING HYBRIDIZATION Detecting natural hybridization between fishes is often complicated because several situations are possible. Two closely related species, or conspecific populations, might occur in the same habitat without ever interbreeding, yet hybridization may be suggested because of phenotypic overlap. On the other hand, hybridization might occur, but the hybrids themselves may never breed for one of several reasons, including infertility. If hybrids backcross with one or both paternal species, introgression may have occurred or a hybrid swarm may be present. If hybridization is known to have occurred, one may want to distinguish individuals of mixed ancestry from those of the two parental species. However, as will be pointed out later, distinguishing individuals of mixed ancestry from parental species is often impossible if hybridization has proceeded past the F1 generation. Morphology Hubbs & Kuronuma (1942) and Hubbs et al (1943) defined a statistic called the hybrid index in order to measure the average morphological similarity of an individual fish to each of the two species or other taxa. The index (I) is calculated separately for each character as I = 100 x [(u - X)/(Y-X)], where u is the value of the trait for the individual being evaluated and X and Y are the mean values of the trait for species X and Y. An individual fish with a value for the trait equal to X or Y will have an index value equal to 0 or 100, respectively. An index value of 50 indicates exact intermediacy for the character in question. The average value of the index for all diagnostic characters may be close to 50 in both known and suspected hybrids, but the individual values may vary widely from character to character (e. g. Ross & Cavender 1981). Multivarite methods Two major objections to the hybrid index are that (1) it requires the a priori indentification of individuals from the two parental species and (2) it fails to account for the variances and covariances of the discriminating traits. Highly correlated traits are often different measurements of the same biological phenomena, reflecting the pleiotropic action of genes or the common response to environmental effects (Falconer 1981). Hubb's hybrid index has lost ground to multivariate statistical methods. Multivariate statistical methods can circumvent these objections by deriving weighting factors for each trait based on the variance-covariance structure of the data set. Discriminant function analysis (DFA) derives weighted linear functions of the measured traits so that two or more groups of individuals are maximally separated or discriminated in multivariate space. DFA also requires a priori classification, but exclusion and inclusion probabilities for each indiviudal are given. But the function can be distorted if two many individuals are at first, falsely classified. Thus, the a priori classification should be done based on information independant of the variables used in the function. Principal components analysis (PCA) is a second multivariate statistical method that is often used in hybridization studies. Unlike DFA, PCA does not require the a priori identification of groups or individuals. PCA only derives a new set of uncorrelated variables, the principal components, which are simple linear transformations of the original variables. An example of a PCA plot (first vs. second component), based on the morphology of two hybridizing fish taxa PCA is relatively easy to apply, and is an excellent exploratory technique. In this example involving a species that we work with (Brachymystax lenok), 37 morphological characters were used. While this researcher (Sergey Alekseyev) did preclassify individuals (therefore the circles are colored), it is clear that he would not have had to. Also, his technique is obviously good, as he was also able to predict the hybrids, which, appear to be F1‘s, though no genetic data is used here! PCA does not, however, provide probabilities of any classification, but the components can be used in subsequent analyses as input variables into a large array of other statistical techniques. Here is another example of PCA used in the analysis of hybridization. Instead of a bivariate plot, the first principal component is simply plotted against the frequency of individuals. Such a technique can be informative when many of the original traits (or at least those that are important) are correlated, and thus much of the variation in the data set is contained within the first component. Figure 7.1 Frequency distributions of first principal component scores for two cyprinid fishes, Notropis cornutus and N. chrysocephalus, and their suspected hybrids from each of five geographic locations. From Dowling and Moore (1984). Common shiner Notropis cornutus Here is an example from our own work, with grayling in Asia. After phenotypic examination, as well as the application of mtDNA typing, the question arose as to whether two sympatric populations were really reproductively isolated. Examination of morphological characters in multvariate space show complete distinction of the „Orange-spot“ and „Lower Amur“ forms. Note that neither PCA factor alone (nor any of the original variables) show nonoverlapping ranges, but considered together, there is clear separation. The additional application of 10 nulcear markers (microsatellites) confirmed that there was no evidence of any hybridization between these two forms, and thus, they could for instance fulfill the BSC criteria of two distinct species. Note as well, the morphological position of the third group (Upper Amur) which is allopatric to the other two. What mechanism did you hear of in the last lecture that might explain the morphological relationship of these three groups? So you can see why the study of hybridization (i.e. hybrid zones) is closely linked to the study of speciation. Hybird zones are model systems, where many of the mechanisms of reproductive isolation can be investigated. The discovery of hybrids alone, however, especially if they are only F1‘s , has little meaning beyond recognizing that a prezygotic barrier is not existent. Post-zygotic mechanisms can be fully responsible (and often are) for preserving the identity of two popuation groups (or species) where hybridization is frequent. Prezygotic mechanisms may be more prone to breaking down under a changing environment (or through anthropogenic influence) and thus the frequency of such mechansims in freshwater fishes might parellel the frequency with which hybridization and introgression is observed. One additional scenario, common in some groups of freshwater fishes, is worthy of demonstrating. Similar to the three-spine stickleback, parallel „speciation“ as been proposed to occur among sympatric forms of many salmonid fishes, exemplified by the genus Coregonus (found also in Salzkammergut!) Sympatric dwarf and normal whitefish ecotypes (Coregonus sp.) Sympatric dwarf and normal whitefish ecotypes (Coregonus sp.) Such sympatric forms are often found in post-glacial lake systems (that is, relatively young systems) whereby not only are F1 hybrids found, but substantial degrees of introgression (i.e seen at genetic markers). But the forms are real, and little phenotypic overlap is found. What mechanism can maintain the existence of two such extreme forms in the presence of substantial gene flow? Detecting hybridization by genetic methods Detecting natural hybridization and introgression by genetic methods is relatively straightforward when the two parental species are completely fixed for different alleles at one or more loci. Individuals of the two parental species will each be homozygous for different alleles, whereas F1 hybrids will be heterozygous for these alleles at all diagnostic loci. A DIAGNOSTIC LOCUS Species 1 Locus 1 F1 Hybrid Species 2 A/A (all individuals) a/a (all individuals) A/a If hybridization has proceeded past the F1 stage, however, hybrid descendants will express a range of different genotypes, including the two parental types. When populations are not „fixed“ for alternate alleles, the problem is more complicated, and individuals can not be assigned to hybrid or pure classes. Thus, hybridization indices must be quantitative. To quantitatively demonstrate the likelihood of hybridization, Campton & Utter (1985) devised a hybrid index measuring the relative probability that the composite genotype for each fish arose by random mating within each of the two species. The index (IH) was defined as: IH = 1.0 - log10 (px) / log10 (px) + log10 (py), where px and py are the conditional probabilities that the composite genotype at all loci for an individual could have arisen by random mating within species X and species Y, respectively, given the average allele frequencies for the two species and assuming gametic phase (linkage) equilibrium among all loci. The power of genetic methods to detect natural hybridization will increase as the number of distinquishing loci increases. This is easily demonstrated by the following table. Number of distinguishing loci Probability of heterozygosity if one allele is found at 0.8 and the other at 0.2 across 1 to 6 different loci Population 1 2 3 4 5 6 Parental (P) 0.3200 0.1024 0.0328 0.0105 0.0034 0.0011 F1 Hybrid (Ph) 0.6800 0.4624 0.3144 0.2138 0.1454 0.0989 43.3 92.1 Increased probability of a heterozygous hybrid Ph/P 2.13 4.52 9.60 20.4 If the frequencies of alternate alleles (e.g. A and a) at a locus are 0.8 and 0.2 in one population and 0.2 and 0.8 in the second population, the probability of an individual being heterozygous in each population is simply 2pq = 0.32. If the two populations interbreed, then the expected frequency of heterozygotes among the F1 hybrids is 0.68, which is only 2.13 times the expected frequency of heterozygotes in each of the parental populations. As the number of distinguishing loci increase, however, this ratio also increases. For example, with four distinguishing loci, the frequency of fourlocus heterozygous among F1 hybrids is more than 20 times their expected frequency in each of the parental populations (Table 7.1). This ratio increases to 92.1 with six distinguishing loci. Thus, even without diagnostic markers, strong inferences about hybridization can be made, when a number of loci are used. For more hypervariable markers (like microsatellites), increasingly sophisticated statistical techniques are available for estimating degrees of introgression (termed admixture) even when allele frequencies of the parent populations are not known. However, there are also prior assumptions and tradeoffs depending on the situation. The more information, the better. Some aspect of the pedigree history is usually needed to make reliable inferences, (number of generations since contact, prior alelle frequencies, number of hybrid categories, etc.). Anderson EC & Thompson (2002) A method for indentifying species hybrids using multilocus genetic data. Genetics 160, 1217-1229. and other references therein..... Mitochondrial DNA – mtDNA can be an extremely helpful tool in understanding the dynamics of hybridization and introgression, but only when applied in combination with nuclear (i.e. bi-parentally inherited markers)! That is, mtDNA helps to identify the source of the mother in a hybrid situation, but alone, mtDNA can not distinguish between current hybridization and ancient processes. mitochondrial capture, or nuclear replacement, a consequence of introgressive hybridization X nDNA Y mtDNA nDNA X mtDNA imagine a male of one species (X), crossing with a female of another (Y); if we consider the black fill to represent the genome of the female, both for nuclear and mtDNA, what will the F1 generation look like in terms of the proportion of each parental donor? nDNA mtDNA The F1 generation will have 100% mtDNA of the female parent (Y), and a 50/50 mixture of both parents (X + Y) for the nuclear genome. in the next generation, a male X crosses with a hybrid female (a backcross) nDNA mtDNA nDNA mtDNA X what happens to the proportions of the parent genomes now? the resulting backcross offspring still retains 100% mtDNA from species Y, but now possesses only 25% of its nuclear genome. With each similar backcross, approximately ½ of the remaining nuclear genome of species Y will be lost and the result can quickly be the complete nuclear „replacement“ or „apparent“ mitochondrial capture of Y‘s mtDNA into a population of X‘s nuclear gene pool Taxa X nDNA Taxa Y mtDNA nDNA mtDNA nDNA mtDNA X nDNA nDNA mtDNA nDNA mtDNA ? mtDNA Thus, a small amount of interspecific gene flow permits transmission of one species‘ mtDNA genome into another species. Several mechanisms can accelerate the rate of such an event. First, an adaptive mutation in the mtDNA genome of the female parent can lead to a „selective sweep“ and ultimate fixation of one haplotype in two hybridizing populations. Second, population bottlenecks during glaciation and resulting genetic drift could accelerate the rate of mtDNA replacement. Such events, like hybridization in general, can be promoted through environmental change (which changes the selection landscape), anthropogenic actions such as the transport and stocking of one species or strain, or any reason that reduces the density of one of the species, promoting the chance mating with another. Studies using only mtDNA, can easily be misleading, and such events may have occured a long time ago, obscuring or confounding phylogenetic relationships. My first tiger, isn‘t she pretty! This is a photo of a so-called „tiger trout“, which is a cross between the brown trout (Salmo trutta) and brook trout (Salvelinus fontinalis). Such crosses are produced in captivity, but the fertility of such organisms in the wild is not well understood. The caption, came withthe photo, and demonstrates the novelty of such hybrids among fisherman. This cross exists in the wild in Austria, and I have evidence of introgession into wild populations of brown trout in the Tirol. Brook trout are introduced in Austria, but also readily hybridize with native populations of Arctic charr (Salvelinus alpinnis).