* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Class Notes 2
Crystallographic database wikipedia , lookup
Halogen bond wikipedia , lookup
Chemical biology wikipedia , lookup
Bent's rule wikipedia , lookup
Physical organic chemistry wikipedia , lookup
Hydrogen-bond catalysis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Western blot wikipedia , lookup
Bond valence method wikipedia , lookup
Crystal structure wikipedia , lookup
History of molecular theory wikipedia , lookup
Hypervalent molecule wikipedia , lookup
Self-assembling peptide wikipedia , lookup
Chemical bond wikipedia , lookup
Structural integrity and failure wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Bottromycin wikipedia , lookup
Hydrogen bond wikipedia , lookup
Protein adsorption wikipedia , lookup
Resonance (chemistry) wikipedia , lookup
Metalloprotein wikipedia , lookup
Biochemistry wikipedia , lookup
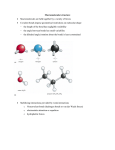
Protein backbone • Biochemical view: – basic repeating unit is NH—CαH—C’=O • We can also look at repeating units from Cα to Cα – Interesting properties: • Bond lengths almost equal in all groups, in all proteins • Bond angles almost equal in all groups, in all proteins • A Cα atom belongs to two units • All atoms in an unit coplanar – Preferable when describing structural properties Protein backbone • Geometric/Structural view: polypeptide chain divided into – Peptide units • • • • • Cα atom and carboxyl group of residue i Amino group and Cα atom of residue i+1 Are rigid groups Rotation on bond C-N is prevented by energy barrier Peptide units are joined by covalent bonds between Cα atoms. Thus – Peptides can rotate along 2 bonds: • N-Cα and Cα-C – Two dihedral angles for each unit: Ф (Phi) and Ψ (Psi) • Two degrees of freedom per unit • Determine the conformation of the backbone Dihedral angles and regular structures • Repeating values of Ф and Ψ along the main chain result in regular structure – repeating values of Ф =-57o and Ψ =-47o give a right-handed helical fold (α-helix) – repetitive values of Ф[-110,-140] and Ψ[+110,+135] give sub chains with conformations that allow interactions between nearby parallel segments (β-sheet) • Most combinations of Ф and Ψ angles are not allowed – Allowed conformations plotted as 2-D chart • Ramachandran plot Secondary Structure • defined by patterns of hydrogen bonds between backbone amide groups – sidechain-mainchain and sidechain-sidechain hydrogen bonds are irrelevant • The amino acids in the interior/core of a globular protein have hydrophobic side chains – Water soluble proteins fold to pack hydrophobic side chain into interior – Results in hydrophobic core and hydrophilic surface – The main chain must fold into interior, too • Main chain is hydrophilic: – – – • N--H: hydrogen bond donor C=O: hydrogen bond acceptor These groups must be neutralized by formation of H bonds secondary structure Secondary structure – α-helices – β-sheets – form rigid and stable frameworks α-Helix • Righthanded coiled conformation – backbone N-H group i+4 forms hydrogen bonding with backbone C = O group i – 3.6 residues per turn (5.4 Å, 1.5 Å per residue) • Variations, with chain more loosely or tightly coiled are possible (i+3 or i+5 instead of i+4) but not often – backbone (φ, ψ) dihedral angles around (-60o,-45o) – Sum of φ and ψ angles of consecutive residues about 105o • Has between 4 to 40 residues • All H bonds point in the same direction – Aligned along helical axis – Dipole moments for residues are aligned along axis • Net dipole for α-helix (+ at N-H end and – at C=O end) Representations (cartoon, backbone trace, space filling) β-sheets • Combination of several regions of the chain (not chain adjacent): β-strands – Parallel: all amino acids go in same direction • Evenly spaced H bonds – Antiparallel: amino acids in successive strands alternate directions • Alternate narrowly/widely spaced H bonds – Mixed β-sheet also exist • • Have twisted strands: right-handed twist (always) β-strand: 5 to 10 residues long – Almost fully extended Representations (bond, cartoon, ribbon) From secondary structure to structure • Protein structure: built from secondary structures – Connected by loop regions • • • • Various lengths Irregular shape Are at the surface of the protein Reach in charged and polar residues – Easier to predict! • In homologous proteins almost always insertions and deletions occur in the loop regions. Structure Motifs • Secondary structures connected to form motifs – α-helices and β-sheets in a motif • Adjacent in the 3-dimensional structure • Connected bu loop regions • Combinations of motifs and secondary structures domains • Tertiary structure: – Arrangement of secondary structure – Structural domains • Quaternary structure – More than one polypeptide folded together • Native conformation: direct consequence of – primary structure – chemical environment • water based • oily interior of a cell membrane – So far, no reliable computational method exists to predict the native structure from the amino acid sequence Structure Classes • Protein structure four classes: – α-domains • core built up only from α-helices – β-domains • core built up only from (usually 2) antiparallel β-sheets – α/β-domains • mostly β-α-β motifs – (mostly) parallel β-sheets surrounded by α-helices – α+β-domains (few cases) • antiparallel β-sheet packed against α-helices