* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Cladistics - Cougar science rocks!
Survey
Document related concepts
Transcript
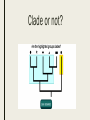
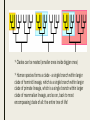
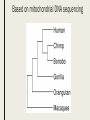
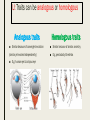
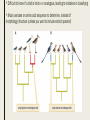
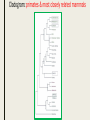
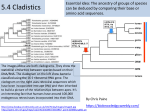
CLADISTICS Topic 5.4 U: A clade is a group of organisms that have evolved from a common ancestor Clade = group of organisms that includes a single ancestor & all of its descendants = unbroken lines of evolutionary descent Includes: ■ Common ancestor ■ Evolved species now extinct ■ Current species - can be 10,000 (birds) or just 1 (Ginkgo biloba) Clade or not? * Clades can be nested (smaller ones inside bigger ones) * Human species forms a clade - a single branch within larger clade of hominid lineage, which is a single branch within larger clade of primate lineage, which is a single branch within larger clade of mammalian lineage, and so on, back to most encompassing clade of all: the entire tree of life! U: Evidence for which species are part of a clade can be obtained from base sequences of a gene or corresponding amino acid sequence of a protein ■ If common ancestor is recent, then few differences in base or amino acid sequence ■ If common ancestor is distant (millions of ya), then many differences in base or amino acid sequence U: Sequence differences accumulate gradually so there is a positive correlation between number of differences between 2 species & time since they diverged from common ancestor ■ Differences in base sequence are result of mutations ■ Differences accumulate over time ■ Evidence that mutations occur at constant rate, so can be used as a “clock” ■ # of differences correlates to how long ago species split from common ancestor ■ “Molecular Clock” = rate at which mutations occur, giving us a timeline Based on mitochondrial DNA sequencing U: Traits can be analogous or homologous Analogous traits Homologous traits ■ Similar because of convergent evolution ■ Similar because of similar ancestry (similar yet evolved independently) ■ E.g. pentadactyl forelimbs ■ E.g. human eye & octopus eye * Difficult to know if a trait is homo- or analogous, leading to mistakes in classifying * Must use base or amino acid sequence to determine, instead of morphology/structure (unless you want to include extinct species!) U: Cladograms are tree diagrams that show the most probable sequence divergence in clades ■ Cladogram = tree diagram based on similarities & differences between species in a clade ■ Based on base or amino acid sequences ■ Principle of parsimony = the simplest explanation is usually the correct one http://science.howstuffworks.com/science-vs-myth/31651conspiracy-test-occams-razor-video.htm ■ “An ancestor of mine maintained that when you eliminate the impossible, whatever remains, however improbable, must be the truth.” -Spock (Star Trek) ■ Clades show the most probable sequence of divergence cladograms ■ Nodes = branching points (represent hypothetical ancestor) ■ Usually 2 clades branch off at a node, but sometimes more ■ Where do these Pterosaurs fit in? App: Cladograms including humans & other primates ■ Closest relatives of humans are chimpanzees & bonobos ■ Entire genomes have been sequenced and compared ■ Molecular clock with mutation rate of 10-9 yr-1 ■ Primates = mammals that have adaptations for climbing trees (humans, monkeys, baboons, gibbons, lemurs) Primate cladogram Cladogram: primates & most closely related mammals Skill: analysis of cladograms to deduce evolutionary relationships (DBQ p273) Answers: U: Evidence from cladistics has shown that classifications of some groups based on structure did not correspond with the evolutionary origins of a group of species ■ Cladistics = construction of cladograms and identification of clades ■ Base and amino acid sequencing is relatively new (end of 20th century) ■ As a result, some groups have been reclassified, revealing unnoticed similarities and differences between species NOS: falsification of theories: Plant families have been reclassified as a result of evidence from cladistics. ■ Important process in science: testing of theories and replacement of theories found to be false with new theories ■ Example: reclassification of plants based on discoveries in cladistics (like Figworts) ■ Classification of angiospermophytes into families based on morphology by French botanist Antoine Laurent de Jussieu in 1789 ■ Revised repeatedly App: Reclassification of the figwort family using evidence from cladistics ■ Until recently, Scrophulariaceae (figwort) was 8th largest family of angiosperm (out of 400+ families) ■ Due to cladistics discoveries, 5 clades had incorrectly been combined into one family ■ Some families were merged, some genera were moved to another family ■ Less than half of species stayed in family ■ Figwort family now 36th largest family