* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Nucleic acids as biomarkers Sequence Conservation Level
Horizontal gene transfer wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Human microbiota wikipedia , lookup
Bacterial morphological plasticity wikipedia , lookup
Marine microorganism wikipedia , lookup
Triclocarban wikipedia , lookup
Metagenomics wikipedia , lookup
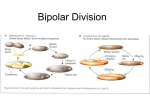
Diversity of Soil Microbes Approaches for Assessing Diversity Microbial community Culture Organism isolation Phenotype Nucleic acid extraction Molecular characterization Nucleic acids as biomarkers 16S rRNA Sequence Conservation Leve nearly universal intermediate hypervariable Phylogenetic categories of organisms Comparative analysis of 16S ribosomal RNA genes Ancient Taxa: The Kingdoms Remotely Related Taxa: The Classes and Divisions (Phyla) QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Not shown are candidate divisions, organisms detected by PCR-16S analysis, but no currently isolated and cultured representative Moderately Related Bacterial Taxa: The Major Intradivisional Groupings (Orders and Families) QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Highly Related Taxa: The Genera and Species Example: Phylogeny of Ammonia-oxidizing bacteria Organisms of the same species are >97% identical in 16S rRNA gene. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Reptitive element (REP)-PCR Genomic fingerprinting 16S rRNA analysis = changes in one gene (DNA sequence) at one site in the genome REP = short sequences that are occur in multiple locations throughout the bacterial genome REP-PCR assays variation in sequence at multiple sites throughout the genome Patterns differentiate bacteria at subspecies level Genomic DNA denatured to expose repetitive sequences Primers bind the repetitive sequences DNA sequences between repetitive sequences are amplified DNA fragments of various lengths are generated www.bacbarcodes.com Fragments are resolved by gel electrophoresis yielding complex fragment patterns Similar strains show similar patterns REP-PCR Analysis of fluorescent Pseudomonas isolates from different sites and continents Fluorescent Pseudomonas: Produce bright pigments in culture Commoly isolated from soils and plant rhizospheres Metabolically diverse Are strains globally mixed or endemic? Examine REP-PCR patterns of bacteria isolated from different continents, and different sites within continents. REP-PCR Analysis of fluorescent Pseudomonas isolates from different continents Genotypes of isolates from different locations form separate clusters Indicates genotypes were endemic QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Summary: Geographic distribution •Fluorescent pseudomonads and others are globallydistributed •Development of endemic species occurs at finer scales •Spatial (geographic) diversification isolation affects bacterial Effects of plant growth on microbial community composition How do plants alter the composition of soil microbial communities? Are certain bacteria preferentially selected for colonization of the rhizosphere? Do certain plants select for certain bacteria? What has a greater impact on the composition of plant rhizospheres? The plant type or the soil? Rhizosphere: General effects Density (abundance) of bacteria increases in rhizosphere relative to bulk soil Bacterial diversity in rhizosphere decreases relative to soil. Increase in abundance of Proteobacteria relative to other phyla Comparison of rhizosphere vs. soil effects Different populations of pseudomonads colonize the rhizosphere of the same plants grown in different soils Unique to Soil 1 Unique to Soil 2 Rhizosphere Effects Growth and enrichment in plant rhizosphere Sub-populations selected for growth from each soil Selection imposed by rhizosphere varies with plant, and plants (root) age Soil 1 Population Soil 2 Population Rhizosphere Effects Plant selects for organisms from a pool that has developed and established in the soil. Different parts of the pool may be selected by different plants Since plant changes with time, selection also varies with time for a given plant Rhizosphere variability over time QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. DGGE profiles of Pseudomonas rhizosphere communities at the early (A) and late (B) flowering stage and at the senescent growth stage (C). Different lanes (1–3) represent rhizosphere sample from different pots. GM, transgenic plants without herbicide; G, transgenic plants with Basta application; MM, wild-type plant without herbicide application; B, wild-type plants with Butisan S application. Bands 2a, 2b, 2c and 2d had the same mobilities as bands 1a (3a), 3b, 1f and 1d. FEMS Microbiol. Ecol. 41:181-190 Perterbations: General effects on soil microbial communities Perterbations: Typically decrease diversity, select for a proliferation of subpopulations Examples: Plants- rhizosphere Animals - Earthworm casts Geological - volcanic eruption Climatic - Fire Anthropogenic - Pollution, agricultural practices Do bacterial community structure changes (decreased diversity) induced by perturbations affect function? Transformations A B C D Group 1 Group 2 Group 3 Group 4 Group 5 Group 6 Functions possesed by the indicated group Stability in transformation of A -->D is supported by diversity in organisms and establishment of redundancy in the activities they possess. How is decreased diversity (less redundacy) reflected in community activity? Community Dilution Experiment Hypothesis: Reduced diversity, reduces redundancy, and reduces the capability of soils to respond to stress Approach: Sterile soil inoculated with serial dilution of soil suspension Incubated 9 months Measured biomass, assessed community diversity, and activities Dilution experiment results: Biomass and diversity No significant difference in biomass, but Decreasing diversity as function of dilution Quic kTime™ and a TIFF (Uncompres sed) dec ompres sor are needed to see this picture. The overall biodiversity in sterile soil inoculated with dilutions of a soil suspension (A, 10 0; B, 10 2; C, 10 4; D, 10 6). The number of species within the individual populations measured (i.e. soil bacterial DNA bands, cultivable bacterial morphotypes, cultivable fungal morphotypes and protozoan species) was normalized relative to the maximum number observed, summed and divided by four (i.e. the number of taxonomic groups) to give the biodiversity index. The bar represents ± one standard error, n=3 Dilution experiment results: Molecular analysis of bacterial diversity Decreasing number of bands, decreasing diversity Quic kTime™ and a TIFF (Uncompres sed) dec ompres sor are needed to see this picture. The DNA banding pattern of soil bacteria obtained by DGGE analysis of eubacterial-primer based amplicons from sterile soil inoculated with dilutions of a soil suspension (A, 100; B, 102; C, 104; D, 106). The three lanes of each treatment represent individual replicates, n=3 Dilution experiment results: Activity as a function of diversity Quic kTime™ and a TIFF (Uncompres sed) dec ompres sor are needed to see this picture. Effect of stresses, in the form of Cu addition or heat treatment, on the ability of sterile soil inoculated with dilutions of a soil suspension (A, 10 0; B, 10 2; C, 10 4; D, 10 6) to decompose grass residues at increasing time intervals following the application of the stress. Bars represent one standard error, n=3. Community Dilution Experiment-Conclusion Diversity could be experimentally manipulated No detectable change in activity with the level of diversity reduction achieved. Minimum level of diversity (functional redundancy) to support process stability in soil is unknown.