* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Characterization of Human Immunodeficiency Virus Type 1 from a
Survey
Document related concepts
Transcript
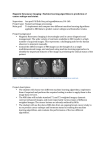
5407_13_p103-109 1/12/05 10:19 AM Page 103 AIDS RESEARCH AND HUMAN RETROVIRUSES Volume 21, Number 1, 2005, pp. 103–109 © Mary Ann Liebert, Inc. Sequence Note Characterization of Human Immunodeficiency Virus Type 1 from a Previously Unexplored Region of South Africa with a High HIV Prevalence PASCAL OBONG BESSONG,1 CHIKWELU LARRY OBI,1 TONIE CILLIERS,2 ISAAC CHOGE,2 MARY PHOSWA,2 CANDICE PILLAY,2 MARIA PAPATHANASOPOULOS,2 and LYNN MORRIS2 ABSTRACT HIV prevalence in the Limpopo Province has increased rapidly within the past 10 years, as in other parts of South Africa. Little is known about the genetic and biological properties of HIV circulating in this region including the baseline drug resistance profiles. We therefore collected blood samples from 42 HIV-1-infected patients residing in this region for analysis. All samples were shown to belong to HIV-1 subtype C by env and gag heteroduplex mobility assay (HMA). Viral isolates from 14 of these patients were shown to use the CCR5 coreceptor exclusively and had gp120 V3 loop sequences consistent with this phenotype. Sequence analysis of both protease and reverse transcriptase genes showed that none of 13 isolates harbored primary resistance mutations. These data suggest that HIV-1 subtype C is the predominant subtype circulating in the Limpopo Province, and that viral strains from this region are indistinguishable from those found in other parts of South Africa. INTRODUCTION T HIV-1 IN SOUTH AFRICA has been among the fastest in the world (www.unaids.org). An antenatal survey conducted in 2001 showed that 24.5% of pregnant women were HIV infected.1 Prevalence rates varied among the nine provinces with KwaZulu-Natal (33.5%), Free State (30.1%), Gauteng (29.8%), and Mpumalanga (29.2%) recording the highest levels. The Limpopo Province has experienced a sharp rise in HIV seroprevalence over the past 10 years, from 0.64% in 1992 to 14.5% in 2001.1,2 It is one of the poorest provinces, largely rural, and highly dependent on the migrant labor system. Its geographic location makes it the portal of entry for surrounding countries of Botswana, Mozambique, and Zimbabwe, which also have high rates of HIV infection. While there have been a number of studies on HIV-1 genetic diversity in South Africa, none has specifically addressed the genetic subtypes of HIV-1 in the Limpopo Province. HE RATE OF SPREAD OF The vast majority of the estimated 4.7 million infected South Africans are infected with HIV-1 subtype C viruses. At an earlier stage of the epidemic, subtype B viruses were identified among homosexual men who reported contacts in the United States.3 Subtypes A, D, CRFO1-AE, and other recombinant viruses have also been identified, although in relatively smaller numbers.3–7 Thus, while subtype C viruses dominate the epidemic, the exceptionally high prevalence rates in South Africa could provide significant opportunities for the spread of new genetic subtypes and/or evolution of current subtypes. Analysis should therefore focus on previously unexplored regions where the opportunity for the introduction of new variants is high and where prevalence rates are rising, such as the Limpopo Province. Given the high prevalence rates in South Africa, there is a desperate need for the introduction of antiretroviral (ARV) therapies. While ARV use is currently restricted to mother-to-child prevention programs, more widespread use is expected in the 1Department 2AIDS of Microbiology, University of Venda, Thohoyandou, South Africa. Virus Research Unit, National Institute for Communicable Diseases, Johannesburg, South Africa. 103 5407_13_p103-109 1/12/05 10:19 AM Page 104 104 BESSONG ET AL. near future as drug prices decrease and the demand for ARV access in developing countries grows. There is no information regarding baseline drug resistance patterns from the Limpopo Province, which would be important to establish prior to the introduction of ARV treatment programs. This study was therefore carried out to identify circulating HIV-1 subtypes and to analyze genotypic drug resistance profiles in a cross-sectional drug-naive population residing in the Limpopo Province of South Africa. MATERIALS AND METHODS Study population and sample collection Whole blood samples in ethylenediaminetetraacetic acid (EDTA) were collected from 42 HIV-1 seropositive volunteers who were outpatients at the Bela-Bela Clinic in Bela Bela (formerly Warmbaths) in the Limpopo Province of South Africa. Samples were collected between July and August 2001. All patients provided signed informed consent and each completed a questionnaire for the provision of demographic data. CD4 counts were performed when possible at the NICD using a FACSCount (Becton Dickinson, CA). Peripheral blood mononuclear cells (PBMC) were isolated using Ficoll–Hypaque (Amersham Pharmacia, Sweden) gradient centrifugation and stored in liquid nitrogen. Plasma was stored at 70°C. Ethical clearance for this study was obtained from the Research Committee of the University of Venda, South Africa. Viral RNA extraction and RT-PCR for env and gag HIV-1 regions Viral RNA was extracted from 140 l plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA). The V3–V5 region of env was amplified in a nested polymerase chain reaction (PCR) as described.8 A region in p24/p7 of gag was amplified in a nested PCR as described,9 except that the annealing temperature was increased to 55°C. Reverse transcription (RT) was done with AMV RT (Roche Diagnostics) at 42°C for 60 min for both PCR reactions. Amplification was carried out in a GeneAmp 9700 thermocycler (Applied Biosystems, Perkin Elmer) for 35 cycles, for the first and second PCR. Negative controls were included in all PCR reactions. Reference plasmids A1 (92RW20, Rwanda), B1 (BR20, Brazil), B2 (TH14, Thailand), C1 (MA959, Malawi), C2 (ZM18, Zambia), D1 (UG21, Uganda), and E1 (TH22, Thailand) were amplified in env and A1 (VI310, Rwanda), B2 (UG280, Uganda), C3 (UG286, Uganda), C4 (ZM145, Zambia), and D1 (K31, Kenya) were amplified in gag. Primers and plasmids were provided in the HIV-1 env and HIV-1 gag heteroduplex mobility assay (HMA) subtyping kits obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Viral phenotypes and V3 sequencing HIV-1 isolation by PBMC coculture was attempted in a subset of patients. Viral phenotype was determined in MT-2 cells as previously described.10 Coreceptor usage was monitored in U87.CD4.CCR5 and U.87.CD4.CXCR4 cell lines with a p24 antigen readout (DuPont NEN, Life Sciences, Boston, MA). Cultures with increasing levels of p24 antigen and evidence of syncytium formation up to day 12 were considered positive for viral replication. The amino acid sequences of the env V3 region of cultured viral isolates was determined as described.10 Drug resistance genotyping RT and PR gene regions were amplified from primary virus isolates as described for the RT11 and PR (Candice Pillay, National Institute for Communicable Diseases, Johannesburg, South Africa, unpublished). Sequences were edited using Sequencher v4.0.5 software and the predicted amino acids were translated using DNasis software. Nucleotide sequences were submitted to the Stanford database for query (HIV-SEQ and beta test) for the detection of mutations associated with drug resistance and closest subtype identity (www.hivdb.stanford.edu/hiv). Phylogenetic analysis The PR and RT nucleotide sequences were aligned using ClustalX with HIV-1 subtypes A–J obtained from the Los Alamos Sequence Database (http://hiv-web.lanl.gov). Additional subtype C PR and RT sequences from Botswana (n 8), Zimbabwe (n 10), and South Africa (n 20) were included.11–16 Dendograms were created by the neighbor-joining method with a bootstrapping stringency of 1000. Comparisons of nucleotide homology among the PR and RT nucleotide sequences were done according to the Kimura two-parameter model. RESULTS Characteristics of study population Blood samples from 42 HIV-1-infected individuals attending an outpatient clinic in the Limpopo Province were used in this study. The median age of the group was 34 years (range 20–53). There were 33 females and 9 males; 83% were single and all except one were unemployed. All the subjects were South Africans and reported heterosexual activity. One individual believed he had been infected in Zimbabwe (PB01). Individuals presented with fungal skin infections or abscesses (n 8), pulmonary tuberculosis (n 8), sexually transmitted infections (n 5), diarrhea (n 2), herpes zoster (n 1), and either minor or no conditions (n 18). The median CD4 count for 17 of the 42 (41%) individuals was 334 cells/l (range of 77–1903) with four having a CD4 count below 200 cells/l. None of the individuals had received antiretroviral therapy. Genetic subtypes Heteroduplex formation and mobility analyses HMAs for env and gag were performed as described.6,8,9 DNA heteroduplexes were resolved on a 5% acrylamide gel for 2.5 hr (env) or 2 hr (gag) at 250 V. Plasma samples from all 42 individuals were used in an env and gag PCR. Amplification products were obtained for 39 (93%) samples in env and 36 (85%) samples in gag. Two samples (01PB14ZA and 01PB23ZA) did not amplify in env and 5407_13_p103-109 1/12/05 10:19 AM Page 105 SOUTH AFRICAN HIV-1 SUBTYPE C five samples (01PB11ZA, 01PB20ZA, 01PB37ZA, 01PB38ZA, and 01PB40ZA) did not amplify in gag. For one sample (01PB19ZA), amplification was not successful for either gene. HMA profiles indicated that all samples belonged to HIV-1 subtype C. No ambiguous or indeterminate profiles were obtained and no other subtype or intersubtype recombinants were identified. All profiles were unique, indicating no evidence for contamination. For the gag HMA, the heteroduplexes migrated closer to the homoduplexes compared to the env HMA, confirming the lower level of genetic diversity in gag. In some cases there was insufficient discrimination between references A and C and it was therefore considered important to include more than one subtype C reference for the gag HMA. An example is shown in Figure 1 (01PB17ZA), where heteroduplexes with references C4 and A1 migrated similarly but reference C3 clearly distinguished this sample as a subtype C. MT-2 coreceptor usage, and V3 loop sequences Virus isolation from PBMC was successful for 14 of 32 (44%) of individuals. All viruses were nonsyncytium inducing (NSI) in an MT-2 cell assay suggestive of CCR5 usage. Indeed all viruses caused extensive syncytia in the U87.CD4.CCR5 cell line with high p24 antigen levels (4.3–250 ng/ml) (Table 1). No syncytia formation was observed in the U87.CD4.CXCR4 cell line and cultures remained p24 antigen negative after 12 days. Analysis of the V3 loop predicted amino acid sequences of 10 105 isolates (sequence data of 01PB12ZA, 01PB13ZA, 01PB22ZA, and 01PB23ZA could not be reliably interpreted) revealed conservation of the GPGQ motif, characteristic of subtype C viruses. All were 35 amino acids in length with a neutral amino acid at position 11 and a negative or neutral amino acid at position 25 and an overall charge of between 2 and 5, typical of R5 isolates (Fig. 2). Drug resistance mutations The PR and RT genes of the 14 viral isolates were analyzed to determine if any harbored drug resistance mutations. One isolate did not amplify in PR (01PB12ZA) and one did not amplify in RT (01PB15ZA). The Stanford sequence database listed all viruses as subtype C in both PR and RT regions. The predicted amino acid PR sequences relative to a subtype C consensus are shown in Figure 3A. There were no primary resistance mutations. Polymorphisms occurred at sites associated with secondary drug resistance and included K20R (6/13), I36L (1/13), L63P/T/V (7/13), V77I (2/13), V82I (1/13), and L93I (1/13). M36I and I93L, which are known secondary drug resistance mutations in subtype B infections, occurred in 12 of 13 (93%) of these subtype C isolates. The predicted amino acid sequences of 13 RT genes are shown in Figure 3B. No primary resistance mutations were detected, but polymorphisms at drug resistance sites included A98S (2/13), V118I (3/13), V179I (3/13), and L210V (1/13). FIG. 1. A representative profile of gag (A) and env (B) HMA analyses. Mobilities of heteroduplexes formed between the amplified gag product of 01PB17ZA and reference strains. The fastest mobility is exhibited by the heteroduplex between the unknown 01PB17ZA and the reference strains C3 (Uganda). Lane S is the sample hybridized unto itself. (B) The fastest heteroduplex is equally shown between sample 01PB17ZA and a reference strain C2 (ZM18, Zambia). Sample 01PB17ZA was hence designated HIV-1 subtype C based on the gag and env regions. 5407_13_p103-109 1/12/05 10:19 AM Page 106 106 BESSONG ET AL. TABLE 1. CORECEPTOR USAGE AND MT-2 PHENOTYPES OF ISOLATES p24 values (ng/ml) and syncytium formation on day 8 Isolate U.87.CD4.CCR5 Syncytia U.87.CD4.CXCR4 Syncytia MT-2 Biotype 12.4 25.3 218.0 53.0 12.0 251.0 179.0 17.3 75.4 4.3 11.6 91.8 5.6 147.2 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.21 0.05 0.05 0.05 NSIa NSI NSI NSI NSI NSI NSI NSI NSI NSI NSI NSI NSI NSI R5 R5 R5 R5 R5 R5 R5 R5 R5 R5 R5 R5 R5 R5 01PB01ZA 01PB02ZA 01PB03ZA 01PB04ZA 01PB08ZA 01PB12ZA 01PB13ZA 01PB15ZA 01PB17ZA 01PB18ZAb 01PB21ZA 01PB22ZA 01PB23ZA 01PB27ZA aNSI, bData nonsyncytium inducing. are from day 12 cultures. Phylogenetic analysis Phylogenetic analysis of the RT nucleotide sequences with reference sequences from different subtypes confirmed that all isolates clustered with subtype C (Fig. 4). A similar phylogenetic relationship was obtained with the PR sequences (data not shown). Pairwise nucleotide distance analysis indicated a 2.1–10.2% variation for the PR and 2.4–7.2% for the RT regions. Comparison of RT sequences with those from other geographic locations in southern Africa including Botswana (n 8), Zimbabwe (n 10), and South Africa (10 from Durban and 10 from Johannesburg) revealed intermixing of these subtype C sequences. This suggests that viruses from the Limpopo Province are indistinguishable from viruses circulating in other parts of southern Africa. DISCUSSION Most of the work performed to date on genetic diversity of HIV in South Africa has focused on urban centers or highly populated regions.3,6,17,18 The Limpopo Province is the poorest and most rural and one of the least studied of the nine provinces of South Africa. In selecting an area for study, we chose Bela-Bela, which is a holiday resort with luxury hotels and a thriving sex industry. All 42 individuals residing in this region were infected with HIV-1 subtype C strains that were indistinguishable from other sequences from the southern African region. In determining the HIV-1 subtypes we employed the heteroduplex mobility assay. This assay has been shown to be a FIG. 2. V3 loop amino acid sequences, MT-2 phenotypes, and coreceptor biotypes of 10 South African HIV-1 subtype C isolates. The sequences were all 35 amino acids in length, with overall charges between 2 and 5. The tetrapeptide motif GPGQ (shown in the box) characteristic of nonsyncytium-inducing HIV-1 subtype C viruses was conserved in all the isolates. Positions 11 and 25, known to determine coreceptor phenotype, are shaded in gray. 5407_13_p103-109 1/12/05 10:19 AM Page 107 SOUTH AFRICAN HIV-1 SUBTYPE C 107 A B FIG. 3. Predicted amino acid sequences of protease (A) and reverse transcriptase (B) of 13 drug-naive South African individuals. The sequences were aligned against a consensus generated from the sequences. There were no deletions or insertions. The dots indicate identical amino acids and changed amino acids are shown. Shaded regions represent drug-resistance-related loci. Polymorphisms at these positions are shown in boldface. rapid, inexpensive, and reliable technique for screening HIV-1 genetic subtypes, especially when both gag and env HMA are used.6,8,19,20 Using this approach we found that all samples in this cohort were HIV-1 subtype C, with no other subtype or intersubtype identified. Phylogenetic analyses of the PR and RT nucleotide sequences provided additional information on their subtype C designation. It is therefore unlikely that these viruses were intersubtype recombinants, although this possibility cannot be excluded based on this subgenomic analysis. All the viruses tested made use of the CCR5 coreceptor and did not cause syncytia in the MT-2 cell line, irrespective of CD4 count or the presence of an AIDS-defining illness. This is in agreement with previous reports in which subtype C viruses were shown to preferentially use CCR5 and rarely induce syncytia.10,21 Our findings on subtype distribution are consistent with those from studies conducted in other regions and show that HIV-1 subtype C is the major subtype in the expanding HIV-1 epidemic in South Africa.6,22 Examination of the protease and reverse transcriptase genes did not reveal any primary resistance mutations to any RT or PR in- 5407_13_p103-109 1/12/05 10:20 AM Page 108 108 BESSONG ET AL. FIG. 4. Phylogenetic relationship of RT sequences. RT sequences from the Limpopo Province (in bold) do not form a separate cluster, but intermingle with subtype C sequences from Johannesburg (00ZA.AC01–00ZA.AC12), Durban (ZA004p01– ZA130p01), Botswana (AF110967, AF110980, AF443115–AF443110), and Zimbabwe (U83614–U83605). hibitors. However, polymorphisms associated with secondary drug resistance among subtype B viruses were detected in subtype C PR sequences. M36I and I93L associated with resistance to each of the PIs occurred in 12 of 13 (92%) sequences while K20R associated with resistance to indinavir, ritonavir, and lopinavir occurred in 6 of 13 (46%) of the patients studied. V82I, which confers low level resistance to nelfinavir, was found in 1 of 13 (8%) sequences. Among the RT sequences, 3 of 13 (23%) viruses had V118I, which confers resistance to nucleoside RTIs when present with E44A/D. Two viruses (15%) had the A98S mutation, while V179I was detected in three viruses (23%). The frequencies of A98S, V118I, and V179I in this cohort were marginally higher than that reported by Pillay et al.,11 in which 1 (0.03%), 3 (0.08%), and 2 (0.05%) of A98S, V118I, and V179I, 5407_13_p103-109 1/12/05 10:20 AM Page 109 SOUTH AFRICAN HIV-1 SUBTYPE C respectively, were observed among 37 pregnant women attending antenatal clinics in Gauteng Province, South Africa. Based on these genotypic profiles, it is expected these viruses would be susceptible to the currently available PIs and RTIs. However, the impact of secondary resistance mutations and polymorphisms on the propensity to develop resistance once drug pressure is applied is unknown. There is therefore a need for continuous surveillance among patients failing antiretroviral therapies in South Africa to determine the resistance patterns among subtype C viruses. This study, to the best of our knowledge, is the first on the molecular epidemiology and genotypic resistance profiles of HIV-1 in the Limpopo Province of South Africa. The results on subtyping may have relevance for the development and evaluation of candidate AIDS vaccines for South Africa. The drug resistance data may be useful in the formulation and implementation of drug delivery policies and programs in the Limpopo Province of South Africa. SEQUENCE DATA The sequence data for the protease, reverse transcriptase, and envelope V3 loop gene regions obtained in this study have been submitted to GenBank under the following accession numbers: AY510031–AY510043, AY510044–AY510056, and AY510057– AY510066, respectively. 109 8. 9. 10. 11. 12. 13. 14. 15. ACKNOWLEDGMENTS 16. This study received funding from the Fogarty International Centre (TWO-0231) and the National Research Foundation, South Africa. We thank Ms. Rebecca Baloyi of the Bela-Bela Clinic for recruitment of volunteers and sample collection and Ms. Mia Coetzer and Mr. Shayne Loubser of NICD for assistance with HMA and phylogenetic analyses, respectively. L.M. is a Wellcome Trust Overseas Senior Research Fellow in Biomedical Sciences. Research done by P.O. Bessong was in partial fulfillment for a Ph.D. degree in the Department of Microbiology, University of Venda, South Africa. REFERENCES 1. Department of Health: National HIV and syphilis seroprevalence survey of women attending public antenatal clinics in South Africa, 2001. 2. Kustner HG, Swanevelder JP, and Van Middelkoop A: National HIV surveillance—South Africa, 1990–1992. S Afr J Sci 1994; 84:195–200. 3. Williamson C, Engelbrecht S, Lambrick M, et al.: HIV-1 subtypes in different risk groups in South Africa. Lancet 1995;346:782. 4. Becker ML, De Jager G, and Becker WB: Analysis of partial gag and env gene sequences of HIV type 1 strains from South Africa. AIDS Res Hum Retroviruses 1995;11:1265–1267. 5. Van Harmelen J, Wood R, Lambrick M, Rybicki EP, and Williamson C: An association between HIV-1 subtypes and mode of transmission in Cape Town, South Africa. AIDS 1997;11:81–87. 6. Bredell H, Hunt Gillian, Morgan B, Tiemessen CT, Martin DJ, and Morris L: Identification of HIV type 1 intersubtype recombinants in South Africa using env and gag heteroduplex mobility assays. AIDS Res Hum Retroviruses 2000;16:493–497. 7. Papathanasopoulos M, Cilliers T, Morris L, et al.: Full-length genome analysis of HIV-1 subtype C utilizing CXCR4 and intersub- 17. 18. 19. 20. 21. 22. type recombinants isolated in South Africa. AIDS Res Hum Retroviruses 2002;18:879–886. Delwart EL, Herring B, Rodrigo AG, and Mullins J: Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl 1995;4:S202–S216. Heyndrickx L, Janssens W, Zekeng L, et al.: Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J Virol 2000;74:363–370. Morris L, Ciliers T, Bredell H, Phoswa M, and Martin DJ: CCR5 is a major coreceptor used by HIV-1 subtype C isolates from patients with active tuberculosis. AIDS Res Hum Retroviruses 2001;17:697–701. Pillay C, Bredell H, McIntyre J, Gray G, and Morris L: HIV-1 subtype C reverse transcriptase sequences from drug naive pregnant women in South Africa. AIDS Res Hum Retroviruses 2002;18:605–610. Shafer RW, Eisen JA, Merigan TC, and Katzenstein D: Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol 1997;71:5441–5448. Shafer RW, Chuang TK, Hsu P, White CB, and Katzenstein DA: Sequence and drug susceptibility of subtype C protease from human immunodeficiency virus type 1 seroconverters in Zimbabwe. AIDS Res Hum Retroviruses 1999;15:65–69. Novitsky VA, Montano MA, McLane MF, et al.: Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: A set of 23 full-length clones from Botswana. J Virol 1999;75:4427–4432. Novitsky V, Smith UR, Gilbert P, et al.: Human immunodeficiency virus type 1 subtype C molecular phylogeny: Consensus sequence for an AIDS vaccine design? J Virol 2002;76:5435–5451. Gordon M, De Oliveira T, Bishop K, et al.: Molecular characteristics of human immunodeficiency virus type 1 subtype C viruses from KwaZulu-Natal, South Africa: Implications for vaccine and antiretroviral control strategies. J Virol 2003;77:2587–2599. Engelbrecht S, Laten JD, Smith TL, and Van Rensburg EJ: Identification of envelope subtypes in fourteen HIV type 1 isolates from South Africa. AIDS Res Hum Retroviruses 1995;11:1269–1271. Bredell H, Williamson C, Sonnenberg P, Martin DJ, and Morris L: Genetic characterization of HIV type 1 from migrant workers in three South African gold mines. AIDS Res Hum Retroviruses 1998;14:677–684. Loussert-Ajaka I, Menu E, Apetrei C, et al.: HIV type 1 diversity and the reliability of the heteroduplex mobility assay. AIDS Res Hum Retroviruses 1998;14:877–883. Tatt ID, Barlow KL, and Clewley JP: A gag gene heteroduplex mobility assay for subtyping HIV-1. J Virol Methods 2000; 87:41–51. Peeters M, Vincent R, Perret J-L, et al.: Evidence for differences in MT-2 cell tropism according to genetic subtypes of HIV-1: Syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol 1999;20:115–121. Engelbrecht S, Smith TL, Kasper P, et al.: HIV type 1 V3 domain serotyping and genotyping in Gauteng, Mpumalanga, KwaZuluNatal and Western Cape Provinces of South Africa. AIDS Res Hum Retroviruses 1999;15:325–328. Address reprint requests to: Lynn Morris AIDS Virus Research Unit National Institute for Communicable Diseases Private Bag X4 Sandringham 2131 Johannesburg, South Africa E-mail: [email protected]