* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Developing a diagnostic service for Stargardt disease – a feasibility
Survey
Document related concepts
Transcript
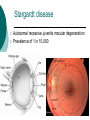
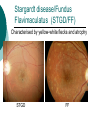
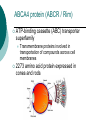
Developing a diagnostic service for Stargardt disease – a feasibility study Emily Packham Oxford Regional Molecular Genetics Laboratory Introduction Inherited eye disorders Services currently available for some of the AD and X-linked conditions Limited services currently for AR conditions (Asper Ophthalmics offer commercial genotyping of some genes) Why? Significant clinical overlap Genetically heterogeneous Stargardt disease may be feasible Stargardt disease Autosomal recessive juvenile macular degeneration Prevalence of 1 in 10,000 Stargardt disease/Fundus Flavimaculatus (STGD/FF) Characterised by yellow-white flecks and atrophy STGD FF Symptoms Age of onset varies from early childhood to twenties Early stages – difficulty reading, watching TV, missing patches in vision, photophobia, slow dark adaption Later stages – always disturbance of central vision and sometimes: peripheral disturbance, increasing photophobia or problems with dark vision Diagnosis Clinical diagnosis Sophisticated imaging but dependent on tests performed, experience and stage of disorder Late stage shows clinical overlap Genetic diagnosis Support or confirm diagnosis Provide prognosis information Aid genetic counselling Therapeutic intervention Genetics of Stargardts disease ABCA4 (1p13-p22) 50 exons (6819bp ORF) Highly polymorphic No mutation hotspots 500+ variants identified Most common seen in ~ <10% of patients Many missense variants ABCA4 protein (ABCR / Rim) ATP-binding cassette (ABC) transporter superfamily Transmembrane proteins involved in transportation of compounds across cell membranes 2273 amino acid protein expressed in cones and rods ABCA4 function and disease pathology Actively ‘flips’ Ret-PE across disc rim membrane Enables retinal signalling to continue Loss-of-function mutations Loss of/reduction in ABCA4 function results in accumulation of toxic lipofuscin deposits Destroys retinal pigment epithelium and rod and cone cells, resulting in visual loss ABCA4 and other retinopathies Stargardt disease AR cone-rod dystrophy AR retinitis pigmentosa Age-related macular degeneration? Genotype/phenotype correlation model based on residual activity of protein Screening strategy 30 patients selected for testing Highly polymorphic, 50 exon gene with no particular hotspots Bi-directional sequencing Robotics approach –5 patients per batch Pathogenicity investigations performed MLPA Results 37 different potential pathogenic variants detected in 26 patients No. of variants No. of patients 0 4 1 6 2 18 3 2 13/20 patients with two or more variants had all of them classified as either likely or highly likely Most common seen in 4 patients Results Results Variant classification Number detected Highly likely pathogenic 17 Likely pathogenic 10 Intermediate 10 Total 37 Extensive published data MLPA normal in all 10 patients tested Feasibility Clinical sensitivity 67% or 43% (+/- intermediate variants) Higher than literature Different screening methods and patient selection Clinical utility Able to interpret most variants Supports clinical diagnosis, aids counselling and therapy Improves equity of access What next? Report our 30 patients Determine if variants are in trans Submit gene dossier Collaborate with BRC retinal research project Evaluating use of high throughput sequencing to test numerous inherited retinal conditions NHS lab BRC Acknowledgements Oxford Molecular Genetics Laboratory Oxford Clinical Genetics and BRC Anneke Seller Treena Cranston Tina Bedenham, Louise Williams, Kate Gibson Andrea Nemeth Oxford eye hospital Susan Downes