* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download The Grand Challenge in Metagenomics Sensitive and
Human microbiota wikipedia , lookup
Bacterial cell structure wikipedia , lookup
Bacterial morphological plasticity wikipedia , lookup
Plant virus wikipedia , lookup
Introduction to viruses wikipedia , lookup
Microorganism wikipedia , lookup
Triclocarban wikipedia , lookup
Community fingerprinting wikipedia , lookup
Phospholipid-derived fatty acids wikipedia , lookup
History of virology wikipedia , lookup
Metagenomics wikipedia , lookup
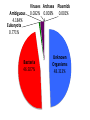
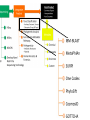
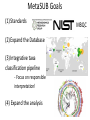
The Grand Challenge in Metagenomics Sensitive and Accurate Taxa Classification Ebrahim Afshinnekoo MetaSUB Global Project Director Mason Lab Weill Cornell Medical College June 20th, 2015 @nycpathomap Ambiguous 4.184% Eukaryota 0.771% Viruses Archaea Plasmids 0.032% 0.003% 0.001% Bacteria 46.927% Unknown Organisms 48.313% 0.003% 0.001% Unknown Organisms 48.313% But what about everything else…? Ambiguous 4.184% Eukaryota 0.771% Viruses Archaea Plasmids 0.032% 0.003% 0.001% Bacteria 46.927% Unknown Organisms 48.313% We’re only as good as our databases… …and there is a great need for standards! Nucleic Acids Research Group More coming from the Metagenomics Research Group –stay tuned! lu En s_ m te eg ro St ba at ap er ct iu hy er m _a lo co er cc og u en s_ Rh es e od pi de os pi rm Ps ril eu id lu is do m _r m on ub as ru _a m er M ug icr in oc os o St cc a re us pt _l om ut eu yc s es Kl _g eb ris sie eu l s l a_ Sp ox or yt os oc ar a cin a_ ur Ba ea ci l En e lu te s _c ro er co eu cc s us _f ae ca lis Ba cil % of MegaBLAST Reads / Expected 30% 25% BLAST Expected 5% 0% 20 MetaPhlAn 20% 15 15% 10 10% 5 0 MetaPhlAn Abundance 35% 25 Comparing the tools… 0.4 0.35 0.3 0.25 0.2 Expected BLAST 0.15 MetaPhlAn OneCodex 0.1 GOTTCHA CosmosID 0.05 0 The “Holy Grail” in Metagenomics One Tool to rule them all One Tool to find the taxa One Tool to bring relative abundances And in the metagenomics bind them A more integrative approach… http://microbe.net/2015/02/17/the-long-road-from-data-to-wisdom-and-from-dna-to-pathogen/ B. anthracis found by Kraken, MetaPhlAn, SURPI, BWA, PhyloSift. But scant pXO, no PlcR SNP. Thus, no evidence of a pathogen in the subway. http://read-lab-confederation.github.io/nyc-subway-anthrax-study/ https://github.com/poeli/GOTTCHA Great name, sounds good, I wonder what it’ll find in some samples… Conclusion: The “G” in GOTTCHA actually stands for Gonorrhea MetaSUB Goals (1) Standards (2) Expand the Database (3) Integrative taxa classification pipeline - Focus on responsible interpretation! (4) Expand the analysis MBQC Thanks to the Swabbing Teams! www.pathomap.org/people/