* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Part 6
Survey
Document related concepts
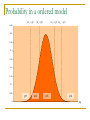
Transcript
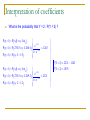
SKEMA Ph.D programme 2010-2011 Class 6 Qualitative Dependent Variable Models Lionel Nesta Observatoire Français des Conjonctures Economiques [email protected] Structure of the class 1. The linear probability model 2. Maximum likelihood estimations 3. Binary logit models and some other models 4. Multinomial models 5. Ordered multinomial models 6. Count data models The Linear Probability Model The linear probability model When the dependent variable is binary (0/1, for example, Y=1 if the firm innovates, 0 otherwise), OLS is called the linear probability model. Y 0 1x1 2 x 2 u How should one interpret βj? Provided that OLS4 – E(u|X)=0 – holds true, then: E(Y | X) 0 1x1 2 x 2 The linear probability model Y follows a Bernoulli distribution with expected value P. This model is called the linear probability model because its expected value, conditional on X, and written E(Y|X), can be interpreted as the conditional probability of the occurrence of Y given values of X. E(Y | X) Pr(Y 1| X) 1 E(Y | X) Pr(Y 0 | X) β measures the variation of the probability of success for a one-unit variation of X (ΔX=1) E(Y | X) Pr(Y 1| X) Pr(Y 1| X) X X Limits of the linear probability model (1) Non normality of errors OLS6 : The error term is independent of all RHS and follows a normal distribution with zero mean and variance σ² u Normal(0, ) 2 Since the errors are the complement to unity of the conditional probability, they follow a Bernoulli distribution, not a normal distribution. Limits of the linear probability model (1) 1.5 1 .5 0 Density 2 2.5 Non normality of errors -1 -.5 0 Residuals .5 Limits of the linear probability model (2) Heteroskedastic errors OLS5 : The variance of the error term, u, conditional on RHS, is the same for all values of RHS Var u x1 , x 2 , , xk 2 The error term is itself distributed Bernoulli, and its variance depends on X. Hence it is heteroskedastic Var(u) P(1 P) E(Y | X) (1 E(Y | X)) Limits of the linear probability model (2) -1 -.5 Residuals 0 .5 Heteroskedastic errors .4 .6 .8 Fitted values 1 1.2 Limits of the linear probability model (3) Fallacious predictions By definition, a probability is always in the unit interval [0;1] 0 EY | X 1 But OLS does not guarantee this condition Predictions may lie outside the bound [0;1] The marginal effect is constant , since P = E(Y|X) grows linearly with X. This is not very realistic (ex: the probability to give birth conditional on the number of children already born) Limits of the linear probability model (3) 2 3 Fallacious predictions 0 1 Density Fallacious predictions .4 .6 .8 Fitted values 1 1.2 Limits of the linear probability model (4) A downward bias in the coefficient of determination R² Observed values are 1 or 0, whereas predictions should lie between 0 and 1: [0;1]. Comparing predicted with observed variables, the goodness of fit as assessed by the R² is systematically low . Limits of the linear probability model (4) .6 .8 1 Fallacious predictions 0 .2 .4 Fallacious predictions which lower the R2 .4 .6 .8 Fitted values 1 1.2 Limits of the linear probability model (4) 1. Non normality of errors 2. Heteroskedastic errors 3. Fallacious predictions 4. A downward bias in the R² u Normal(0, 2 ) Var u x1 , x 2 , , x k 2 0 EY | X 1 Overcoming the limits of the LPM 1. 2. 3. 4. Non normality of errors Increase sample size Heteroskedastic errors Use robust estimators Fallacious prediction Perform non linear or constrained regressions A downward bias in the R² Do not use it as a measure of goodness of fit Persistent use of LPM Although it has limits, the LPM is still used 1. In the process of data exploration (early stages of the research) 2. It is a good indicator of the marginal effect of the representative observation (at the mean) 3. When dealing with very large samples, least squares can overcome the complications imposed by maximum likelihood techniques. Time of computation Endogeneity and panel data problems The LOGIT Model Probability, odds and logit We need to explain the occurrence of an event: the LHS variable takes two values : y={0;1}. In fact, we need to explain the probability of occurrence of the event, conditional on X: P(Y=y | X) ∈ [0 ; 1]. OLS estimations are not adequate, because predictions can lie outside the interval [0 ; 1]. We need to transform a real number, say z to ∈ ]-∞;+∞[ into P(Y=y | X) ∈ [0 ; 1]. The logistic transformation links a real number z ∈ ]-∞;+∞[ to P(Y=y | X) ∈ [0 ; 1].It is also called the link function The logit link function Let us make sure that the transformation of z lies between 0 and1 z ; e z 0; eZ Z Z 0;1 since e 1 e 1 eZ eZ is called the logit link function Z 1 e The logit model Hence the probability of any event to occur is : eZ P y 1| z 1 eZ 1 1 P y 0 | z 1 P y 1| z 1 Z 1 e 1 eZ But what is z? The odds ratio The odds ratio is defined as the ratio of the probability and its complement. Taking the log yields z. Hence z is the log transform of the odds ratio. P 1 eZ Z e Z 1 P 1 e P ln z 1 P This has two important characteristics : 1. Z ∈ ]-∞;+∞[ and P(Y=1) ∈ [0 ; 1] 2. The probability is not linear in z (The plot linking z with straight line) Probability, odds and logit P(Y=1) Odds p(y=1) 1-p(y=1) 0.01 1/99 0,01 -4,60 0.03 3/97 0,03 -3,48 0.05 5/95 0,05 -2,94 0.20 20/80 0,25 -1,39 0.30 30/70 0,43 -0,85 0.40 40/60 0,67 -0,41 0.50 50/50 1,00 0,00 0.60 60/40 1,50 0,41 0.70 70/30 2,33 0,85 0.80 80/20 4,00 1,39 0.95 95/5 19,0 2,94 0.97 0.99 97/3 99/1 32,3 99,0 3,48 4,60 Ln (odds) The logit transformation The preceding table matches levels of probability with the odds ratio. The probability varies between 0 and 1, The odds varies between 0 and + ∞. The log of the odds varies between – ∞ and + ∞ . Notice that the distribution of the log of the odds is symetrical. .15 .1 0 .05 Density .2 .25 Logistic probability density distribution -10 -5 0 Log (Odds ratio) 5 10 .6 .4 .2 0 P(y=1 | z) .8 1 “The probability is not linear in z” -4 -2 0 z 2 4 The logit link function The whole trick that can overcome the OLS problem is then to posit: z 1x1 k x k z = Xβ eZ e Xβ Hence is rewritten Z 1 e 1 e Xβ But how can we estimate the above equation knowing that we do not observe z? Maximum likelihood estimations OLS can be of much help. We will use Maximum Likelihood Estimation (MLE) instead. MLE is an alternative to OLS. It consists of finding the parameters values which is the most consistent with the data we have. In Statistics, the likelihood is defined as the joint probability to observe a given sample, given the parameters involved in the generating function. One way to distinguish between OLS and MLE is as follows: OLS adapts the model to the data you have : you only have one model derived from your data. MLE instead supposes there is an infinity of models, and chooses the model most likely to explain your data. Likelihood functions Let us assume that you have a sample of n random observations. Let f(yi ) be the probability that yi = 1 or yi = 0. The joint probability to observe jointly n values of yi is given by the likelihood function: n f y1 , y2 ,..., yn f ( yi ) i 1 We need to specify function f(.). It comes from the empirical descrite distribution of an event that can have only two outcome : a success (yi = 1) or a failure (yi = 0). This is the binomial distribution. Hence: 1 n f (yi ) pk (1 p)n k p yi (1 p)1 yi k yi f (yi ) p yi (1 p)1 yi Likelihood functions Knowing p (as the logit), having defined f(.), we come up with the likelihood function: n n i 1 i 1 L y f (yi ) p 1 p 1 yi yi 1 yi yi e 1 L y, z f (yi , z) z z 1 e 1 e i 1 i 1 n z n yi 1 yi e 1 L y, x, f (yi , X, β) Xβ Xβ 1 e 1 e i 1 i 1 n n Xβ Log likelihood (LL) functions The log transform of the likelihood function (the log likelihood) is much easier to manipulate, and is written: n n i 1 i 1 LL y, z yi z ln 1 e z n n i 1 i 1 LL y, x, yi Xβ ln 1 e Xβ n LL y, x, ln 1 e Xβ yi Xβ i 1 Maximum likelihood estimations The LL function can yield an infinity of values for the parameters β. Given the functional form of f(.) and the n observations at hand, which values of parameters β maximize the likelihood of my sample? In other words, what are the most likely values of my unknown parameters β given the sample I have? Maximum likelihood estimations The LL is globally concave and has a maximum. The gradient is used to compute the parameters of interest, and the hessian is used to compute the variance-covariance matrix. LL n yi i x i 0 i 1 ez where i z n 1 e ²LL i 1 i x i xi i 1 However, there is not analytical solutions to this non linear problem. Instead, we rely on a optimization algorithm (Newton-Raphson) You need to imagine that the computer is going to generate all possible values of β, and is going to compute a likelihood value for each (vector of ) values to then choose (the vector of) β such that the likelihood is highest. Example: Binary Dependent Variable We want to explore the factors affecting the probability of being successful innovator (inno = 1): Why? 352 (81.7%) innovate and 79 (18.3%) do not. The odds of carrying out a successful innovation is about 4 against 1 (as 352/79=4.45). The log of the odds is 1.494 (z = 1.494) For the sample (and the population?) of firms the probability of being innovative is four times higher than the probability of NOT being innovative Logistic Regression with STATA Instruction Stata : logit logit y x1 x2 x3 … xk [if] [weight] [, options] Options noconstant : estimates the model without the constant robust : estimates robust variances, also in case of heteroscedasticity if : it allows to select the observations we want to include in the analysis weight : it allows to weight different observations Logistic Regression with STATA Let’s start and run a constant only model logit inno Goodness of fit . . logit inno Iteration 0: log likelihood = -205.30803 Logistic regression Number of obs LR chi2(0) Prob > chi2 Pseudo R2 Log likelihood = -205.30803 inno Coef. _cons 1.494183 Std. Err. .1244955 z 12.00 = = = = 431 0.00 . 0.0000 P>|z| [95% Conf. Interval] 0.000 1.250177 1.73819 Parameter estimates, Standard errors and z values Interpretation of Coefficients What does this simple model tell us ? Remember that we need to use the logit formula to transform the logit into a probability : Xβ e P(Y 1| X) Xβ 1 e Interpretation of Coefficients e1,494 P 0,817 1,494 1 e The constant 1.491 must be interpreted as the log of the odds ratio. Using the logit link function, the average probability to innovate is dis exp(_b[_cons])/(1+exp(_b[_cons])) We find exactly the empirical sample value: 81,7% Interpretation of Coefficients A positive coefficient indicates that the probability of innovation success increases with the corresponding explanatory variable. A negative coefficient implies that the probability to innovate decreases with the corresponding explanatory variable. Warning! One of the problems encountered in interpreting probabilities is their non-linearity: the probabilities do not vary in the same way according to the level of regressors This is the reason why it is normal in practice to calculate the probability of (the event occurring) at the average point of the sample Interpretation of Coefficients Let’s run the more complete model logit inno lrdi lassets spe biotech . logit inno lrdi lassets spe biotech Iteration Iteration Iteration Iteration Iteration 0: 1: 2: 3: 4: log log log log log likelihood likelihood likelihood likelihood likelihood = = = = = -205.30803 -167.71312 -163.57746 -163.45376 -163.45352 Logistic regression Number of obs LR chi2(4) Prob > chi2 Pseudo R2 Log likelihood = -163.45352 inno Coef. lrdi lassets spe biotech _cons .7527497 .997085 .4252844 3.799953 -11.63447 Std. Err. .2110683 .1368534 .4204924 .577509 1.937191 z 3.57 7.29 1.01 6.58 -6.01 P>|z| 0.000 0.000 0.312 0.000 0.000 = = = = 431 83.71 0.0000 0.2039 [95% Conf. Interval] .3390634 .7288574 -.3988654 2.668056 -15.43129 1.166436 1.265313 1.249434 4.93185 -7.837643 Interpretation of Coefficients e -11.630.75rdi0.99lassets0.43spe3.79biotech P 1 e-11.630.75rdi0.99lassets0.43spe3.79biotech Using the sample mean values of rdi, lassets, spe and biotech, we compute the conditional probability : e -11.630.75rdi0.99lassets0.43spe3.79biotech P 1 e-11.630.75rdi0.99lassets0.43spe3.79biotech e1.953 0,8758 1.953 1 e Marginal Effects It is often useful to know the marginal effect of a regressor on the probability that the event occur (innovation) As the probability is a non-linear function of explanatory variables, the change in probability due to a change in one of the explanatory variables is not identical if the other variables are at the average, median or first quartile, etc. level. prvalue provides the predicted probabilities of a logit model (or any other) prvalue prvalue prvalue prvalue prvalue prvalue prvalue , , , , , , x(lassets=10) x(lassets=11) x(lassets=12) x(lassets=10) x(lassets=11) x(lassets=12) rest(mean) rest(mean) rest(mean) rest(median) rest(median) rest(median) Marginal Effects prchange provides the marginal effect of each of the explanatory variables for the majority of the variations of the desired values prchange [varlist] [if] [in range] ,x(variables_and_values) rest(stat) prchange prchange, fromto prchange , fromto x(size=10.5) fromto rest(mean) Goodness of Fit Measures In ML estimations, there is no such measure as the R2 But the log likelihood measure can be used to assess the goodness of fit. But note the following : The higher the number of observations, the lower the joint probability, the more the LL measures goes towards -∞ Given the number of observations, the better the fit, the higher the LL measures (since it is always negative, the closer to zero it is) The philosophy is to compare two models looking at their LL values. One is meant to be the constrained model, the other one is the unconstrained model. Goodness of Fit Measures A model is said to be constrained when the observed set the parameters associated with some variable to zero. A model is said to be unconstrained when the observer release this assumption and allows the parameters associated with some variable to be different from zero. For example, we can compare two models, one with no explanatory variables, one with all our explanatory variables. The one with no explanatory variables implicitly assume that all parameters are equal to zero. Hence it is the constrained model because we (implicitly) constrain the parameters to be nil. The likelihood ratio test (LR test) The most used measure of goodness of fit in ML estimations is the likelihood ratio. The likelihood ratio is the difference between the unconstrained model and the constrained model. This difference is distributed c2. If the difference in the LL values is (no) important, it is because the set of explanatory variables brings in (un)significant information. The null hypothesis H0 is that the model brings no significant information as follows: LR 2ln Lunc ln Lc High LR values will lead the observer to reject hypothesis H0 and accept the alternative hypothesis Ha that the set of explanatory variables does significantly explain the outcome. The McFadden Pseudo 2 R We also use the McFadden Pseudo R2 (1973). Its interpretation is analogous to the OLS R2. However its is biased doward and remain generally low. Le pseudo-R2 also compares The likelihood ratio is the difference between the unconstrained model and the constrained model and is comprised between 0 and 1. Pseudo R 2 MF ln Lc ln Lunc ln Lunc 1 ln Lunc ln Lc Goodness of Fit Measures Constrained model . logit inno Iteration 0: log likelihood = -205.30803 Logistic regression Number of obs LR chi2(0) Prob > chi2 Pseudo R2 Log likelihood = -205.30803 . logit inno Coef. _cons 1.494183 Std. Err. .1244955 z 12.00 = = = = 431 0.00 . 0.0000 P>|z| [95% Conf. Interval] 0.000 1.250177 1.73819 LR 2 ln L unc ln Lc 2 163.5 205.3 83.8 inno lrdi lassets spe biotech, nolog Logistic regression Number of obs LR chi2(4) Prob > chi2 Pseudo R2 Log likelihood = -163.45352 inno Coef. lrdi lassets spe biotech _cons .7527497 .997085 .4252844 3.799953 -11.63447 Std. Err. .2110683 .1368534 .4204924 .577509 1.937191 Unconstrained model z 3.57 7.29 1.01 6.58 -6.01 P>|z| 0.000 0.000 0.312 0.000 0.000 = = = = 431 83.71 0.0000 0.2039 [95% Conf. Interval] .3390634 .7288574 -.3988654 2.668056 -15.43129 1.166436 1.265313 1.249434 4.93185 -7.837643 Ps.R 2MF 1 ln L unc ln Lc 1 163.5 205.3 0.204 Other usage of the LR test The LR test can also be generalized to compare any two models, the unconstrained one being nested in the constrained one. Any variable which is added to a model can be tested for its explanatory power as follows : logit [modèle contraint] est store [nom1] logit [modèle non contraint] est store [nom2] lrtest nom2 nom1 Goodness of Fit Measures . logit inno lrdi lassets spe, nolog Logistic regression Number of obs LR chi2(3) Prob > chi2 Pseudo R2 Log likelihood = -191.84522 inno Coef. lrdi lassets spe _cons .9275668 .3032756 .3739987 -.4703812 Std. Err. .1979951 .0792032 .3800765 .9313494 z 4.68 3.83 0.98 -0.51 P>|z| 0.000 0.000 0.325 0.614 = = = = 431 26.93 0.0000 0.0656 [95% Conf. Interval] .5395037 .1480402 -.3709376 -2.295793 1.31563 .4585111 1.118935 1.35503 . est store model1 . logit inno lrdi lassets spe biotech, nolog Logistic regression Number of obs LR chi2(4) Prob > chi2 Pseudo R2 Log likelihood = -163.45352 inno Coef. lrdi lassets spe biotech _cons .7527497 .997085 .4252844 3.799953 -11.63447 Std. Err. .2110683 .1368534 .4204924 .577509 1.937191 z 3.57 7.29 1.01 6.58 -6.01 P>|z| 0.000 0.000 0.312 0.000 0.000 = = = = 431 83.71 0.0000 0.2039 [95% Conf. Interval] .3390634 .7288574 -.3988654 2.668056 -15.43129 1.166436 1.265313 1.249434 4.93185 -7.837643 . est store model2 . lrtest model2 model1 Likelihood-ratio test (Assumption: model1 nested in model2) LR chi2(1) = Prob > chi2 = 56.78 0.0000 LR test on the added variable (biotech) LR 2 ln L unc ln Lc 2 163.5 191.8 56.8 Quality of predictions Lastly, one can compare the quality of the prediction with the observed outcome variable (dummy variable). One must assume that when the probability is higher than 0.5, then the prediction is that the vent will occur (most likely And then one can compare how good the prediction is as compared with the actual outcome variable. STATA does this for us: estat class Quality of predictions . estat class Logistic model for inno True Classified D ~D Total + - 337 15 51 28 388 43 Total 352 79 431 Classified + if predicted Pr(D) >= .5 True D defined as inno != 0 Sensitivity Specificity Positive predictive value Negative predictive value Pr( +| D) Pr( -|~D) Pr( D| +) Pr(~D| -) 95.74% 35.44% 86.86% 65.12% False False False False Pr( +|~D) Pr( -| D) Pr(~D| +) Pr( D| -) 64.56% 4.26% 13.14% 34.88% + + - rate rate rate rate for for for for true ~D true D classified + classified - Correctly classified 84.69% Other Binary Choice models The Logit model is only one way of modeling binary choice models The Probit model is another way of modeling binary choice models. It is actually more used than logit models and assume a normal distribution (not a logistic one) for the z values. The complementary log-log models is used where the occurrence of the event is very rare, with the distribution of z being asymetric. Other Binary Choice models Probit model Pr(Y 1| X) Xβ z e z2 2 2 dz Xβ Xβ 2 2 e Complementary log-log model Pr(Y 1| X) Xβ 1 exp exp( Xβ) 2 dz Xβ t dz Likelihood functions and Stata commands n Logit : L( y, x, ) i 1 1 yi yi e Xβ 1 f ( yi , xi , ) Xβ Xβ i 1 1 e 1 e n n n i 1 i 1 Probit : L( y, x, ) f ( yi , xi , ) ( Xβ) i 1 ( Xβ) y n n i 1 i 1 1 yi Log-log comp : L( y, x, ) f ( yi , xi , ) 1 exp( exp( Xβ)) i exp( exp( Xβ)) Example logit inno rdi lassets spe pharma probit inno rdi lassets spe pharma cloglog inno rdi lassets spe pharma y 1 yi 0 .1 .2 y .3 .4 Probability Density Functions -4 -2 0 x Probit Transformation Complementary log log Transformation 2 4 Logit Transformation 0 .2 .4 y .6 .8 1 Cumulative Distribution Functions -4 -2 0 x Probit Transformation Complementary log log Transformation 2 4 Logit Transformation Comparison of models Ln(R&D intensity) ln(Assets) Spe BiotechDummy Constant Observations OLS Logit Probit C log-log 0.110 0.752 0.422 354 [3.90]*** [3.57]*** [3.46]*** [3.13]*** 0.125 0.997 0.564 0.493 [8.58]*** [7.29]*** [7.53]*** [7.19]*** 0.056 0.425 0.224 0.151 [1.11] [1.01] [0.98] [0.76] 0.442 3.799 2.120 1.817 [7.49]*** [6.58]*** [6.77]*** [6.51]*** -0.843 -11.634 -6.576 -6.086 [3.91]** [6.01]*** [6.12]*** [6.08]*** 431 431 431 431 Absolute t value in brackets (OLS) z value for other models. * 10%, ** 5%, *** 1% Comparison of marginal effects OLS Logit Probit C log-log Ln(R&D intensity) 0.110 0.082 0.090 0.098 ln(Assets) 0.125 0.110 0.121 0.136 Specialisation 0.056 0.046 0.047 0.042 Biotech Dummy 0.442 0.368 0.374 0.379 For all models logit, probit and cloglog, marginal effects have been computed for a one-unit variation (around the mean) of the variable at stake, holding all other variables at the sample mean values. Multinomial LOGIT Models Multinomial models Let us now focus on the case where the dependent variable has several outcomes (or is multinomial). For example, innovative firms may need to collaborate with other organizations. One can code this type of interactions as follows Collaborate with university (modality 1) Collaborate with large incumbent firms (modality 2) Collaborate with SMEs (modality 3) Do it alone (modality 4) Or, studying firm survival Survival (modality 1) Liquidation (modality 2) Mergers & acquisition (modality 3) Multinomial models One could first perform three logistic regressions as follows : P(Y 1| X) (1) ln (1) (1) 0 1 x1 m xm 1 P(Y 1| X) P(Y 2 | X) (2) ln (2) (2) 0 1 x1 m xm 1 P(Y 2 | X) P(Y 3 | X) (3) ln (3) 0 1 x1 1 P(Y 3 | X) (3) m xm Where 1 = survival, 2 = liquidation, 3 = M&A. 1. Open the file mlogit.dta 2. Estimate for each type of outcome the conditional probability of the event for the representative firm - time (log_time) - size (log labour) - firm age (entry_age) - Spin out (spin_out) - Cohort (cohort_*) The need for multinomial models P(Y 1| X) (1) (1) (1) ln x 0 1 1 m xm 1 P(Y 1| X) P(Y 2 | X) (2) (2) (2) ln x 0 1 1 m xm 1 P(Y 2 | X) P(Y 3 | X) (3) (3) ln 0 1 x1 1 P(Y 3 | X) P(Y 1| X) 0.8771 P(Y 2 | X) 0.0398 P(Y 3 | X) 0.0679 (3) m xm P(Y k | X) k 0.9848 1 Multinomial models First, the sum of all conditional probabilities should add up to unity k PY j | X 1 j0 Second, for k outcomes, we need to estimate (k-1) modality. Hence k PY 0 | X 1 PY j | X jk Multinomial logit models Third, the multinomial model is a simultaneous (as opposed to sequential) estimation model comparing the odds of each modality with respect to all others. With three outcomes, we have: P(Y 1| X) (1|0) (1|0) (1|0) ln x 0 1 1 m xm P(Y 0 | X) P(Y 2 | X) (2|0) (2|0) (2|0) ln x 0 1 1 m xm P(Y 0 | X) P(Y 1| X) (1|2) (1|2) ln 0 1 x1 P(Y 2 | X) (1|2) m xm Multinomial logit models Note that there is redundancy, since : P Y 1| X PY 2 | X P Y 1| X ln ln ln PY 0 | X PY 0 | X P Y 2 | X P Y 1| X PY 2 | X P Y 1| X 1|0 2|0 1|2 ln x ;ln x ;ln x PY 0 | X PY 0 | X P Y 2 | X x 1|0 x 2|0 x 1|2 Fourth, the multinomial logit model estimates (k – 1) outcomes with following constrained: 1|0 2|0 1|2 Multinomial logit models With k outcomes, the probability of occurrence of event j reads: x e ( j|0 ) PY j | X jk x e ( j|0 ) j0 By convention, outcome 0 is the base outcome Multinomial logit models PY j | X Note that x j|j ln ln(1) 0 PY j | X x, j: j|j 0 x e ( j|0 ) PY j | X jk x e ( j|0 ) j0 x e ( j|0 ) PY j | X jk 1 j1 e x( j|0 ) PY 0 | X 1 jk 1 j1 x e ( j|0 ) Binomial logit as multinomial logit Let us rewrite the probability of event that Y=1 P Y 1| X e x 1 e x x e (1|0 ) P Y 1| X x 1 e (1|0 ) x e (1|0 ) (1|0 ) x e x e ( 0|0 ) x e (1|0 ) x e ( k|0 ) k 0,1 The binomial logit binomial is a special case of the multinomial where only two outcomes are being analyzed. Likelihood functions Let us assume that you have a sample of n random observations. Let f(yj ) be the probability that yi = j. The joint probability to observe jointly n values of yj is given by the likelihood function: n f y1 , y2 ,..., yn f ( yi ) i 1 We need to specify function f(.). It comes from the empirical discrete distribution of an event that can have several outcomes. This is the multinomial distribution. Hence: f (y j ) p 0 dYi0 dYi1 1 p p j dYij pk dYik pj jK dYik The maximum likelihood function The maximum likelihood function reads: k dYij L(y) f yi p j i 1 i 1 j1 dYi0 dYij ( j|0) x n n k 1 e ( j|0) L(y) f yi , x i , j k j k ( j|0) ( j|0) x x i 1 i 1 j1 1 e 1 e j1 j1 n n The maximum likelihood function The log transform of the likelihood yields ( j|0 ) x i k n 1 e dyij ln LL(y, x, ( j|0) ) dyi0 ln j k j k x i( j|0 ) x i( j|0 ) i 1 j1 1 e 1 e j 0 j 0 j k xi( j|0 ) k j ( j|0) j k xi( j|0 ) ) ln 1 e dyi x i ln 1 e i 1 j 0 j1 j 0 n LL(y, x, ( j|0) LL(y, x, ( j|0) n k ) dy x i i 1 j1 j i ( j|0) k j k xi( j|0 ) k 1 ln 1 e j1 j 0 Multinomial logit models Stata Instruction : mlogit mlogit y x1 x2 x3 … xk [if] [weight] [, options] Options : noconstant : omits the constant robust : controls for heteroskedasticity if : select observations weight : weights observations Multinomial logit models use mlogit.dta, clear mlogit type_exit log_time log_labour entry_age entry_spin cohort_* Goodness of fit Parameter estimates, Standard errors and z values Base outcome, chosen by STATA, with the highest empirical frequency Interpretation of coefficients The interpretation of coefficients always refer to the base category Does the probability of being boughtout decrease overtime ? No! Relative to survival the probability of being bought-out decrease overtime Interpretation of coefficients The interpretation of coefficients always refer to the base category Is the probability of being bought-out lower for spinoff? No! Relative to survival the probability of being bought-out is lower for spinoff Interpretation of coefficients 1|0 2|0 1|2 2|0 1|0 2|1 Relative to liquidation, the probability of being bought-out is higher for spinoff lincom [boughtout]entry_spin – [death]entry_spin Changing base outcome mcross provides other estimates by changing the base ouctome Mind the new base outcome!! Being bought-out relative to liquidation Relative to liquidation, the probability of being bought-out is higher for spinoff Changing base outcome mcross provides other estimates by changing the base ouctome And we observe the same results as before Independence of irrelevant alternatives - IAA The model assumes that each pair of outcome is independent from all other alternatives. In other words, alternatives are irrelevant. From a statistical viewpoint, this is tantamount to assuming independence of the error terms across pairs of alternatives A simple way to test the IIA property is to estimate the model taking off one modality (called the restrained model), and to compare the parameters with those of the complete model If IIA holds, the parameters should not change significantly If IIA does not hold, the parameters should change significantly Independence of irrelevant alternatives - IAA H0: The IIA property is valid H1: The IIA property is not valid 1 * * ˆ ˆ ˆ ˆ H R C var R var C ˆ R ˆ *C The H statistics (H stands for Hausman) follows a χ² distribution with M degree of freedom (M being the number of parameters) STATA application: the IIA test H0: The IIA property is valid H1: The IIA property is not valid mlogtest, hausman Omitted variable Application de IIA H0: The IIA property is valid H1: The IIA property is not valid mlogtest, hausman We compare the parameters of the model “liquidation relative bought-out” estimated simultaneously with “survival relative to bought-out” avec the parameters of the model “liquidation relative bought-out” estimated without “survival relative to bought-out” Application de IIA H0: The IIA property is valid H1: The IIA property is not valid mlogtest, hausman The conclusion is that outcome survival significantly alters the choice between liquidation and bought-out. In fact for a company, being bought-out must be seen as a way to remain active with a cost of losing control on economic decision, notably investment. Ordered Multinomial LOGIT Models Ordered multinomial models Let us now concentrate on the case where the dependent variable is a discrete integer which indicates an intensity. Opinion surveys make an extensive use on such so-called Likert Scale: Obstacles à l’innovation (échelle de 1 à 5) Intensité de collaboration (échelle de 1 à 5) Enquête de marketing (N’apprécie pas (1) – Apprécie (7)) Note d’étudiants Test d’opinion Etc. Ordered multinomial models Such variables depict a vertical scale – quantitative, so that one can think of them as describing the interval in which an unobserved latent variable y* lies: y 1 si y*n 1 y 2 si 1 y*n 2 y 3 si 2 y*n 3 M y k si 3 y*k where αj are unknown bounds to be estimated. Ordered multinomial models We assume that the latent variable y* is a linear combination of the set of all explanatory variables y*i x i u i where ui follows a cumulative distribution function F(.). The probabilities with each occurrence y (y ≠ y*) are then following the cdf F(.). Let us look at the probability that y = 1 : P(y 1) P y*i 1 P(y 1) P x i u i 1 P(y 1) P u i 1 x i e1 xi P(y 1) 1 x i 1 e1 xi Ordered multinomial models The probability that y = 2 is: P(y 2) P y*i 2 P y*i 1 e2 xi e1 xi P(y 2) 2 x i 1 x i 1 e2 xi 1 e1 xi Altogether we have: P(Y 1) 1 x i P(Y 2) 2 x i 1 x i P(Y 3) 3 x i 2 x i M P(Y k) 1 k 1 x i Probability in a ordered model 1 xi 2 x i 3 xi k1 xi 0.45 0.4 0.35 0.3 0.25 0.2 0.15 0.1 0.05 y=1 0 y=2 y=3 y=k ui The likelihood function The likelihood function is n k L(y, x, , ) = F( j x i) F( j-1 x i) i=1 j=1 avec F( 0 - x n) 0 F( k - x n) 1 dy j The likelihood function If ui follows a logistic distribution, the (log) likelihood function reads : n x x j i j-1 i k e e x x j i j=1 1 e 1 e j-1 i dy j L(y, x, , ) = i=1 et donc x e j x i e j-1 i LL(y, x, , ) = dy ln j x i j-1 x i i 1 j1 1 e 1 e n k j i Ordered multinomial logit models Stata Instruction : ologit ologit y x1 x2 x3 … xk [if] [weight] [, options] Options : noconstant : omits the constant robust : controls for heteroskedasticity if : select observations weight : weights observations Ordered multinomial models use est_var_qual.dta, clear ologit innovativeness size rdi spe biotech Goodness of fit Estimated parameters Cutoff points Interpretation of coefficients A positive (negative) sign indicates a positive relationship between the independent variable and the order (or rank) How does one interpret the cutoff values? The model is: Score x i u i What is then the probability that Y = 1 : P(Y = 1) ? What is the probability that the score be inferior to the first cutoff point? P(y 1) P x i u i 1 e1.95 P(y 1) P 270.5 u i 268.6 .1245 1.95 1 e P(y 1) P u i 1.9 Interpretation of coefficients What is the probability that Y = 2 : P(Y = 2) ? P(y 1) P x i u i 1 e1.95 P(y 1) P 270.5 u i 268.6 .1245 1.95 1 e P(y 1) P u i 1.9 P(y 1) P x i u i 2 e1.95 P(y 1) P 270.5 u i 269.3 .2321 1.95 1 e P(y 1) P u i 1.2 P(Y 2) F 2 x i F 1 x i P(Y 2) .2321 .1245 P(Y 2) .1076 STATA computation of pred. prob. prvalue computes the predicted probabilities Count Data Models Part 1 The Poisson Model Count data models Let us now focus on outcome counting the number of occurrences a given event. Analyzing the number of innovations, the number patents, of invention. Again OLS fails to meet the constrain that the prediction must be nil or positive. To explain count variables, we assume that the dependent variable follows a Poisson distribution. Poisson models Let Y be a random count variabl. The probability that Y be equal to integer yi is given by the Poisson probability density distribution: e i iyi P Y yi , yi 0,1, 2,... yi ! with E Y var Y i To introduce the set of explanatory variables in the model, we condition λi and impose the following log linear form: i e xi ln i x i Poisson distributions 0,5 Lambda 0,45 0,4 0,8 1,5 2,9 10,5 0,35 0,3 0,25 0,2 0,15 0,1 0,05 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 The likehood function The (log) likelihood function reads: ei iyi L(y, ) = yi ! i=1 n et donc LL(y, x, ) = yi x i e x ln yi ! n i i 1 Poisson models Stata Instruction : poisson Poisson y x1 x2 x3 … xk [if] [weight] [, options] Options : noconstant : omits the constant robust : controls for heteroskedasticity if : select observations weight : weights observations Poisson models use est_var_qual.dta, clear Poisson patent lrdi lassets spe biotech Goodness of fit . poisson patent lrdi lassets spe biotech Iteration 0: Iteration 1: Iteration 2: log likelihood = -3549.9316 log likelihood = -3549.8433 log likelihood = -3549.8433 Poisson regression Number of obs LR chi2(4) Prob > chi2 Pseudo R2 Log likelihood = -3549.8433 patent Coef. lrdi lassets spe biotech _cons .699484 .4705428 .7623891 1.073271 -3.12651 Std. Err. .0326467 .0133588 .0441729 .051571 .1971912 z 21.43 35.22 17.26 20.81 -15.86 P>|z| 0.000 0.000 0.000 0.000 0.000 = = = = 431 1766.11 0.0000 0.1992 [95% Conf. Interval] .6354977 .4443601 .6758118 .9721939 -3.512997 Estimated parameters .7634704 .4967256 .8489664 1.174349 -2.740022 Interpretation of coefficients If variables are entered in log, one can interpret the coefficients as elasticities ln x 1 x ln x ; ln i x i x x x patent Coef. lrdi lassets spe biotech _cons .699484 .4705428 .7623891 1.073271 -3.12651 Std. Err. .0326467 .0133588 .0441729 .051571 .1971912 z 21.43 35.22 17.26 20.81 -15.86 P>|z| 0.000 0.000 0.000 0.000 0.000 [95% Conf. Interval] .6354977 .4443601 .6758118 .9721939 -3.512997 .7634704 .4967256 .8489664 1.174349 -2.740022 A one % increase in firm size is associated with a .47% increase in the expected number of patents Interpretation of coefficients If variables are entered in log, one can interpret the coefficients as elasticities ln x 1 x ln x ; ln i x i x x x patent Coef. lrdi lassets spe biotech _cons .699484 .4705428 .7623891 1.073271 -3.12651 Std. Err. .0326467 .0133588 .0441729 .051571 .1971912 z 21.43 35.22 17.26 20.81 -15.86 P>|z| 0.000 0.000 0.000 0.000 0.000 [95% Conf. Interval] .6354977 .4443601 .6758118 .9721939 -3.512997 A one % increase R&D investment is associated with a .69% increase in the expected number of patents .7634704 .4967256 .8489664 1.174349 -2.740022 Interpretation of coefficients If variables are not entered in log, the interpretation changes 100 × (eβ – 1) patent Coef. lrdi lassets spe biotech _cons .699484 .4705428 .7623891 1.073271 -3.12651 Std. Err. .0326467 .0133588 .0441729 .051571 .1971912 z 21.43 35.22 17.26 20.81 -15.86 P>|z| 0.000 0.000 0.000 0.000 0.000 [95% Conf. Interval] .6354977 .4443601 .6758118 .9721939 -3.512997 .7634704 .4967256 .8489664 1.174349 -2.740022 A one-point rise in the degree of specialisation is associated with a 113% increase in the expected number of patents Interpretation of coefficients For dummy variables, the interpretation changes slightly patent Coef. lrdi lassets spe biotech _cons .699484 .4705428 .7623891 1.073271 -3.12651 Std. Err. .0326467 .0133588 .0441729 .051571 .1971912 z 21.43 35.22 17.26 20.81 -15.86 P>|z| 0.000 0.000 0.000 0.000 0.000 [95% Conf. Interval] .6354977 .4443601 .6758118 .9721939 -3.512997 .7634704 .4967256 .8489664 1.174349 -2.740022 Biotechnology firms have an expected number of patents which is 191% higher than pharmaceutical companies. Interpretation of coefficients All variables are very significant … but … Variable Obs Mean patent 431 10.83295 Std. Dev. 17.622 E Y var Y Min Max 0 202 Count Data Models Part 2 Negative Binomial Models Negative binomial models Generally, the Poisson model is not valid, due to the presence of overdispersion in the data. This violates the asumption of equality between the mean and variance if the dependent variable implied by the Poisson model. ln vi ln i ln u i x i i The negative binomial model treats this problem by adding to the log linear form a unobserved heterogeneity term ui: P Y yi eiui i u i yi ! yi Negative binomial models The density of yi is obtained by taking the density of ui : eiui i u i i ui 1 f Y yi | x i g u i du i with g u i e ui yi ! 0 y Assuming that ui is distributed Gamma with mean 1, the density of yi reads: yi i f Y yi | x i yi 1 i i yi Likelihood Functions yi yi i L y, , y 1 i i i i 1 n n LL y, x, yi x i yi ln e i 1 x i ln where α is the overdispersion parameter Negative binomial models Stata Instruction: nbreg nbreg y x1 x2 x3 … xk [if] [weight] [, options] Options : noconstant : omits the constant robust : controls for heteroskedasticity if : select observations weight : weights observations Negative binomial models use est_var_qual.dta, clear nbreg poisson PAT rdi size spe biotech Negative binomial regression Number of obs LR chi2(4) Prob > chi2 Pseudo R2 Dispersion = mean Log likelihood = -1374.339 patent Coef. Std. Err. lrdi lassets spe biotech _cons .7823229 .6167106 .8390795 1.515035 -5.179659 .1121344 .0600372 .1950958 .2384884 .8629357 /lnalpha .323967 alpha 1.382602 z 0.000 0.000 0.000 0.000 0.000 431 122.62 0.0000 0.0427 Goodness of fit [95% Conf. Interval] .5625434 .4990399 .4566988 1.047606 -6.870982 1.002102 .7343813 1.22146 1.982464 -3.488337 .0759342 .1751387 .4727953 .1049868 1.191411 1.604473 Likelihood-ratio test of alpha=0: 6.98 10.27 4.30 6.35 -6.00 P>|z| = = = = chibar2(01) = 4351.01 Prob>=chibar2 = 0.000 Estimated parameters Overdispersion parameter Overdispersion test Interpretation of coefficients If variables are entered in log, one can still interpret the coefficients as elasticities A one % increase in firm size is associated with a .61% increase in the expected number of patents Interpretation of coefficients If variables are entered in log, one can still interpret the coefficients as elasticities A one % increase in R&D investment is associated with a .78% increase in the expected number of patents Interpretation of coefficients If variables are not entered in log, the interpretation changes 100 × (eβ – 1) A one-point rise in the degree of specialisation is associated with a 129% increase in the expected number of patents Interpretation of coefficients For dummy variables, the interpretation follows the same transformation 100 × (eβ – 1) Biotechnology firms have an expected number of patents which is 352% higher than pharmaceutical companies. Overdispersion test We use the LR test to compare the negative binomial model with the Poisson model LR 2 ln L NBREG ln LPRM 2 3055 6110 -1481 - -4536 The results indicate the probability to reject H0 wrongly is almost nil (H0: Alpha=0). Hence there is overdispersion in the data and as a consequence one shopuld use the negative binomial model Larger standard errors and lower z values Variable Poisson NegBin patent lrdi lassets spe biotech _cons 0.699 21.43 0.471 35.22 0.762 17.26 1.073 20.81 -3.127 -15.86 0.782 6.98 0.617 10.27 0.839 4.30 1.515 6.35 -5.180 -6.00 lnalpha 0.324 4.27 _cons Statistics alpha N 431 1.383 431 legend: b/t Extensions ML estimators All models can be extended to a panel context to take full account of unobserved heterogeneity Fixed effect Random effects Heckman models Selection bias Two equations, one on the probability to be observed Survival models Discrete time (complementary log-log, logit) Continuous time (Cox model)