Application of whole genome sequencing to fully characterise
... using the Illumina platform. From the 504 samples received, WGS data was obtained for 470 Campylobacter isolates, comprising 351 from IID1 and 119 from IID2. Of these 416 were C. jejuni and 46 were C. coli. We also obtained WGS data from five C. upsaliensis, one C. fetus, one Arcobacter butzleri and ...
... using the Illumina platform. From the 504 samples received, WGS data was obtained for 470 Campylobacter isolates, comprising 351 from IID1 and 119 from IID2. Of these 416 were C. jejuni and 46 were C. coli. We also obtained WGS data from five C. upsaliensis, one C. fetus, one Arcobacter butzleri and ...
Inferring Host Gene Subnetworks Involved in Viral
... virus), and infers a subnetwork of directed interactions that provides at least one path from every hit to a predicted interface. By providing these paths, we say that the subnetwork plausibly explains or accounts for the viral phenotype observed when each hit is suppressed. Because the background n ...
... virus), and infers a subnetwork of directed interactions that provides at least one path from every hit to a predicted interface. By providing these paths, we say that the subnetwork plausibly explains or accounts for the viral phenotype observed when each hit is suppressed. Because the background n ...
Documenting Your Data Entry Practices
... HOW TO USE THIS WORK SHEET: This work sheet lists all of the criteria that can be used to identify patients eligible for cancer screening, as well as patients who should be excluded from cancer screening. These criteria are based on current cancer screening guidelines and recommendations. For each c ...
... HOW TO USE THIS WORK SHEET: This work sheet lists all of the criteria that can be used to identify patients eligible for cancer screening, as well as patients who should be excluded from cancer screening. These criteria are based on current cancer screening guidelines and recommendations. For each c ...
Approaches to Repeat Finding
... 2. Extend seeds with Hamming distance and edit (Levenshtein) distance to find approximate repeats ...
... 2. Extend seeds with Hamming distance and edit (Levenshtein) distance to find approximate repeats ...
Stability Approach to Regularization Selection (StARS) for High
... b with high probability. In choose one Λ other words, we want to “overselect” instead of “underselect”. Such a choice is motivated by application problems like gene regulatory networks reconstruction, in which we aim to study the interactions of many genes. For these types of studies, we tolerant so ...
... b with high probability. In choose one Λ other words, we want to “overselect” instead of “underselect”. Such a choice is motivated by application problems like gene regulatory networks reconstruction, in which we aim to study the interactions of many genes. For these types of studies, we tolerant so ...
PPTX - Tandy Warnow
... • MetaPhyler, MetaPhlAn, and mOTU are marker-based techniques (but use different marker genes). ...
... • MetaPhyler, MetaPhlAn, and mOTU are marker-based techniques (but use different marker genes). ...
Package 'MatrixEQTL'
... SlicedData object with genotype information. Can be real-valued for linear models and must take at most 3 distinct values for ANOVA unless the number of ANOVA categories is set to a higher number (see useModel parameter). ...
... SlicedData object with genotype information. Can be real-valued for linear models and must take at most 3 distinct values for ANOVA unless the number of ANOVA categories is set to a higher number (see useModel parameter). ...
PPT - Bioinformatics.ca
... • In many cases in biology, the number of features is much larger than the number of samples • Important features may not be represented in the training data • This can result in overfitting – when a classifier discriminates well on its training data, but does not generalise to orthogonally derived ...
... • In many cases in biology, the number of features is much larger than the number of samples • Important features may not be represented in the training data • This can result in overfitting – when a classifier discriminates well on its training data, but does not generalise to orthogonally derived ...
Flexible expressed region analysis for RNA
... modeling of the number of reads that cross these defined features. We previously proposed an alternative statistical model for finding differentially expressed regions (DERs) that first identifies regions that show differential expression signal and then annotates these regions using previously anno ...
... modeling of the number of reads that cross these defined features. We previously proposed an alternative statistical model for finding differentially expressed regions (DERs) that first identifies regions that show differential expression signal and then annotates these regions using previously anno ...
2002-09-12: Segregation Analysis II
... Avoid Contaminating Mating Types Controlled crosses. Select only the appropriate mating types by ascertaining them through their offspring. ...
... Avoid Contaminating Mating Types Controlled crosses. Select only the appropriate mating types by ascertaining them through their offspring. ...
Model-Based Clustering for Expression Data via a Dirichlet Process
... mixture model based clustering of Medvedovic and Sivaganesan (2002) and Medvedovic et al. (2004). Model-based techniques offer advantages over heuristic schemes, such as the ability to assess uncertainty about the resulting clustering and to formally estimate the number of clusters. This chapter des ...
... mixture model based clustering of Medvedovic and Sivaganesan (2002) and Medvedovic et al. (2004). Model-based techniques offer advantages over heuristic schemes, such as the ability to assess uncertainty about the resulting clustering and to formally estimate the number of clusters. This chapter des ...
http://www.gse-journal.org/articles/gse/pdf/1996/06/GSE_0999-193X_1996_28_6_ART0003.pdf
... a single QTL together with additive polygenic and residual variance components. The REML analysis was implemented with a derivative-free algorithm. The method overcomes the shortcomings of the traditional methods of linear regression (eg, Haley et al, 1994; Zeng, 1994) and maximum likelihood (ML) in ...
... a single QTL together with additive polygenic and residual variance components. The REML analysis was implemented with a derivative-free algorithm. The method overcomes the shortcomings of the traditional methods of linear regression (eg, Haley et al, 1994; Zeng, 1994) and maximum likelihood (ML) in ...
Simple Algorithms to Calculate Asymptotic Null Distributions of
... to three genotypes. The null hypothesis of no association is equivalent to no association in the contingency table. Under the alternative hypothesis, if one of the two alleles confers a high risk of the disease, an individual’s risk having the disease increases with the number of risk alleles in the ...
... to three genotypes. The null hypothesis of no association is equivalent to no association in the contingency table. Under the alternative hypothesis, if one of the two alleles confers a high risk of the disease, an individual’s risk having the disease increases with the number of risk alleles in the ...
phylogenetic tree
... All trees will be assumed to be binary (an edge that branches splits into two daughter edges). Each edge of the tree has a certain amount of evolutionary divergence associated to it. We adopt the general term ‘length’, which will be represented by lengthes of edges on figures. A true biological ...
... All trees will be assumed to be binary (an edge that branches splits into two daughter edges). Each edge of the tree has a certain amount of evolutionary divergence associated to it. We adopt the general term ‘length’, which will be represented by lengthes of edges on figures. A true biological ...
STATISTICAL GENETICS `98 Transmission Disequilibrium, Family
... ASPs is to apply a method that correctly accounts for dependencies among sibs when one is testing for linkage disequilibrium (Martin et al. 1997). Whether the TDT method will be more successful than other linkage methods will very likely depend on the population studied (e.g., whether linkage disequ ...
... ASPs is to apply a method that correctly accounts for dependencies among sibs when one is testing for linkage disequilibrium (Martin et al. 1997). Whether the TDT method will be more successful than other linkage methods will very likely depend on the population studied (e.g., whether linkage disequ ...
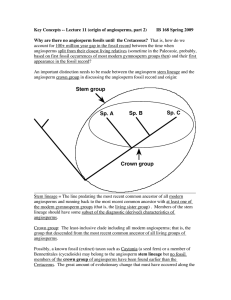
Sp. A Sp. B Sp. C Crown group Stem group
... prior to the origin of modern angiosperms, they were able to root the angiosperm phylogeny without an outgroup, by placing the root between the two gene trees. Less than a month later, Qiu et al. published a paper in Nature based on phylogenetic analysis of 5 genes (from all 3 plant genomes) analysi ...
... prior to the origin of modern angiosperms, they were able to root the angiosperm phylogeny without an outgroup, by placing the root between the two gene trees. Less than a month later, Qiu et al. published a paper in Nature based on phylogenetic analysis of 5 genes (from all 3 plant genomes) analysi ...
Protein Sequence Alignment and Database Searching
... •First scoring matrix was used in the comparison of protein sequences in evolutionary terms by Late Margret Dayhoff and coworkers •Matrices –Dayhoff, MDM, or PAM, BLOSUM etc. •Basic BLAST program does not allow gaps in its alignments •Gapped BLAST and PSI-BLAST ...
... •First scoring matrix was used in the comparison of protein sequences in evolutionary terms by Late Margret Dayhoff and coworkers •Matrices –Dayhoff, MDM, or PAM, BLOSUM etc. •Basic BLAST program does not allow gaps in its alignments •Gapped BLAST and PSI-BLAST ...
The Optimal Discovery Procedure II: Applications to Comparative
... In a microarray study, there is very often pervasive asymmetry in differential expression that is not due to chance. Indeed, it would seem unlikely that overall differential expression would be symmetric, unless the experiment was designed to achieve this behavior. Asymmetric differential expression ...
... In a microarray study, there is very often pervasive asymmetry in differential expression that is not due to chance. Indeed, it would seem unlikely that overall differential expression would be symmetric, unless the experiment was designed to achieve this behavior. Asymmetric differential expression ...
Exploratory data analysis for microarray data
... Metrics and distances A metric d is a function satisfying: 1. non-negativity: d(a, b) ≥ 0; 2. symmetry: d(a, b) = d(b, a); 3. d(a, a) = 0. 4. definiteness: d(a, b) = 0 if and only if a = b; 5. triangle inequality: d(a, b) + d(b, c) ≥ d(a, c). A function only satisfying 1.-3. is called a distance. ...
... Metrics and distances A metric d is a function satisfying: 1. non-negativity: d(a, b) ≥ 0; 2. symmetry: d(a, b) = d(b, a); 3. d(a, a) = 0. 4. definiteness: d(a, b) = 0 if and only if a = b; 5. triangle inequality: d(a, b) + d(b, c) ≥ d(a, c). A function only satisfying 1.-3. is called a distance. ...
lecture4-eQTLmapping
... The genomic regions that contribute to variation in a quantitative phenotype (e.g. blood pressure) ...
... The genomic regions that contribute to variation in a quantitative phenotype (e.g. blood pressure) ...
PartitionFinder manual
... similar models. For example, if you have a dataset of 3 protein-coding genes you might suspect that each of the three genes has been evolving differently – perhaps they come from different chromosomes or have experienced different evolutionary constraints. Furthermore, you might think that each codo ...
... similar models. For example, if you have a dataset of 3 protein-coding genes you might suspect that each of the three genes has been evolving differently – perhaps they come from different chromosomes or have experienced different evolutionary constraints. Furthermore, you might think that each codo ...
Evaluation of the phylogenetic position of the planctomycete
... In recent years, the planctomycetes have been recognized as a phylum of environmentally important bacteria with habitats ranging from soil and freshwater to marine ecosystems. The planctomycetes form an independent phylum within the bacterial domain, whose exact phylogenetic position remains controv ...
... In recent years, the planctomycetes have been recognized as a phylum of environmentally important bacteria with habitats ranging from soil and freshwater to marine ecosystems. The planctomycetes form an independent phylum within the bacterial domain, whose exact phylogenetic position remains controv ...
Detecting epistasis via Markov bases
... has been given in [15]. In the method described in this paper, we first reduce the potential interacting SNPs to a small number by filtering all SNPs genome-wide with a single locus approach. The loci achieving some threshold are then further examined for interactions. Such a two-stage approach has ...
... has been given in [15]. In the method described in this paper, we first reduce the potential interacting SNPs to a small number by filtering all SNPs genome-wide with a single locus approach. The loci achieving some threshold are then further examined for interactions. Such a two-stage approach has ...