Epigenetic memory in mammals
... germline must not only escape reprogramming but also the subsequent wave of de novo methylation that occurs (Morgan et al., 2005). DMRs are crucial to escaping reprogramming and many imprinted genes contain them. DMRs indicate areas that are methylated differently between parental alleles and some, ...
... germline must not only escape reprogramming but also the subsequent wave of de novo methylation that occurs (Morgan et al., 2005). DMRs are crucial to escaping reprogramming and many imprinted genes contain them. DMRs indicate areas that are methylated differently between parental alleles and some, ...
Inheritance of Organelle DNA Sequences in a Citrus–Poncirus
... The apparent segregation of the intergeneric F1 progeny with respect to the P. trifoliata mtDNA configurations suggested that these configurations resulted from influence of the nuclear genome. Nuclear alleles might alter mtDNA organization in the progeny, producing P. trifoliata configurations. Alt ...
... The apparent segregation of the intergeneric F1 progeny with respect to the P. trifoliata mtDNA configurations suggested that these configurations resulted from influence of the nuclear genome. Nuclear alleles might alter mtDNA organization in the progeny, producing P. trifoliata configurations. Alt ...
Prevention of DNA Rereplication Through a Meiotic Recombination
... of Cdk1 at tyrosine 19 (Leu and Roeder 1999). Early work in mitotic cells indicated that Swe1catalyzed Cdk1 phosphorylation regulates the morphogenesis checkpoint (Lew and Reed 1995). However, it is now known that Swe1 is also a component of one of three Mec1-dependent mechanisms that operate in t ...
... of Cdk1 at tyrosine 19 (Leu and Roeder 1999). Early work in mitotic cells indicated that Swe1catalyzed Cdk1 phosphorylation regulates the morphogenesis checkpoint (Lew and Reed 1995). However, it is now known that Swe1 is also a component of one of three Mec1-dependent mechanisms that operate in t ...
Ionic distribution around simple DNA models. I
... out.23 For these reasons, in the context of biophysical studies like drug–DNA or protein–DNA binding, few simulation studies have been conducted at the explicit water level of detail.24 From a physicochemical point of view there is another major shortcoming in using such detailed models. The interpl ...
... out.23 For these reasons, in the context of biophysical studies like drug–DNA or protein–DNA binding, few simulation studies have been conducted at the explicit water level of detail.24 From a physicochemical point of view there is another major shortcoming in using such detailed models. The interpl ...
Regulation of DNA Polymerase Exonucleolytic Proofreading Activity
... proofreading, which removes correct nucleotides in addition to incorrect nucleotides (Muzyczka et al. 1972; Gillin and Nossal, 1976a; reviewed in Goodman et al. 1993). Another potential disadvantage of increased DNA replication accuracy is the possible necessity of a certain minimal mutation rate th ...
... proofreading, which removes correct nucleotides in addition to incorrect nucleotides (Muzyczka et al. 1972; Gillin and Nossal, 1976a; reviewed in Goodman et al. 1993). Another potential disadvantage of increased DNA replication accuracy is the possible necessity of a certain minimal mutation rate th ...
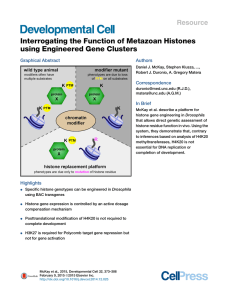
Interrogating the Function of Metazoan Histones using
... to regulation of genome activity by controlling chromatin packaging (Shogren-Knaak et al., 2006), and by serving as binding sites for protein complexes that control a variety of DNA-dependent processes including transcription, replication, and repair (Lachner et al., 2001). Second, histone proteins ...
... to regulation of genome activity by controlling chromatin packaging (Shogren-Knaak et al., 2006), and by serving as binding sites for protein complexes that control a variety of DNA-dependent processes including transcription, replication, and repair (Lachner et al., 2001). Second, histone proteins ...
Method and system for computationally identifying clusters within a
... regulatory regions and additional regions for Which a func tionality has not yet been identi?ed. Protein molecules are synthesiZed from the gene templates in a tWo-step process. In the ?rst step, called transcription, the gene is copied to produce a molecule of messenger ribose-nucleic acid ...
... regulatory regions and additional regions for Which a func tionality has not yet been identi?ed. Protein molecules are synthesiZed from the gene templates in a tWo-step process. In the ?rst step, called transcription, the gene is copied to produce a molecule of messenger ribose-nucleic acid ...
Tissue-preserving approach to extracting DNA from paraffin
... on chromosome arm 18q in 16 individuals. There were no differences in the results obtained using DNA obtained by either technique. In 3 instances in which LOH was detected using the tissue core method, LOH was detected using microdissection as well. Similarly, in all cases in which there was no LOH ...
... on chromosome arm 18q in 16 individuals. There were no differences in the results obtained using DNA obtained by either technique. In 3 instances in which LOH was detected using the tissue core method, LOH was detected using microdissection as well. Similarly, in all cases in which there was no LOH ...
Slides
... 2. Extension causes displacement of strand on other side of nick 3. D-loop is cleaved, invading strand is ligated to newly created 3′-end of the homologous strand 4. 3′-end of newly synthesized strand & the 5′-end of a homologous strand are ligated forming a ...
... 2. Extension causes displacement of strand on other side of nick 3. D-loop is cleaved, invading strand is ligated to newly created 3′-end of the homologous strand 4. 3′-end of newly synthesized strand & the 5′-end of a homologous strand are ligated forming a ...
Comprehensive Analysis of Hyrdrogen Bonds in Regulatory Protein
... of these bonds involve the protein side-chains and the DNA atoms at the base edges and in the backbone, but interactions that involve the protein backbone are also found. The contacts that involve the DNA backbone are believed to stabilize the complex and to orient the protein against the DNA in a f ...
... of these bonds involve the protein side-chains and the DNA atoms at the base edges and in the backbone, but interactions that involve the protein backbone are also found. The contacts that involve the DNA backbone are believed to stabilize the complex and to orient the protein against the DNA in a f ...
File - western undergrad. by the students, for the students.
... Usually, transmembrane segments of proteins have α-helical secondary structure, and if a protein has a single transmembrane segment, it is essentially always an α-helix. The major reason for this is that the α-helix maximizes hydrogen bonding between polar backbone groups, minimizing interactions b ...
... Usually, transmembrane segments of proteins have α-helical secondary structure, and if a protein has a single transmembrane segment, it is essentially always an α-helix. The major reason for this is that the α-helix maximizes hydrogen bonding between polar backbone groups, minimizing interactions b ...
Ionic distribution around simple DNA models. I
... out.23 For these reasons, in the context of biophysical studies like drug–DNA or protein–DNA binding, few simulation studies have been conducted at the explicit water level of detail.24 From a physicochemical point of view there is another major shortcoming in using such detailed models. The interpl ...
... out.23 For these reasons, in the context of biophysical studies like drug–DNA or protein–DNA binding, few simulation studies have been conducted at the explicit water level of detail.24 From a physicochemical point of view there is another major shortcoming in using such detailed models. The interpl ...
pdf
... sulphate (DMS), the DNA will no longer bind to the protein. Thus, DNA is gently methylated (about one hit per molecule), mixed with the protein, and then the bound complexes are separated from the unbound probe. The unbound probe will be modified at all sites (when the whole population of molecules ...
... sulphate (DMS), the DNA will no longer bind to the protein. Thus, DNA is gently methylated (about one hit per molecule), mixed with the protein, and then the bound complexes are separated from the unbound probe. The unbound probe will be modified at all sites (when the whole population of molecules ...
DNA Pre-ConceptionStu - the Biology Scholars Program Wiki
... 11. In order for two strands of DNA to stay together, the two strands must have the same sequence of basic units. A. I’m absolutely certain this is true B. I’m pretty sure this is true C. I have no idea whether this is true or false D. I’m pretty sure it is false E. I’m absolutely certain this is f ...
... 11. In order for two strands of DNA to stay together, the two strands must have the same sequence of basic units. A. I’m absolutely certain this is true B. I’m pretty sure this is true C. I have no idea whether this is true or false D. I’m pretty sure it is false E. I’m absolutely certain this is f ...
Directions and Questions for Lab 9 - San Diego Unified School District
... d. Carefully decant the used stain. Make sure the gel remains flat and does not move up against the corner. Decant the stain directly to a sink drain and flush with water. e. Add distilled or tap water to the staining tray. To accelerate destaining, gently rock the tray. Destain until bands are dist ...
... d. Carefully decant the used stain. Make sure the gel remains flat and does not move up against the corner. Decant the stain directly to a sink drain and flush with water. e. Add distilled or tap water to the staining tray. To accelerate destaining, gently rock the tray. Destain until bands are dist ...
Gene Section FANCD2 (Fanconi anemia, complementation group D2) Atlas of Genetics and Cytogenetics
... in the FA pathway, will then interact with other proteins involved in DNA repair, possibly BRCA1; after DNA repair, FANCD2 return to the nonubiquinated form (FANCD2-S). FANCD2co-localizes with BRCA1 in DNA damagedinduced loci and in the synaptonemal complex of meotic chromosomes as well. ...
... in the FA pathway, will then interact with other proteins involved in DNA repair, possibly BRCA1; after DNA repair, FANCD2 return to the nonubiquinated form (FANCD2-S). FANCD2co-localizes with BRCA1 in DNA damagedinduced loci and in the synaptonemal complex of meotic chromosomes as well. ...
Stage-Specific Histone Modification Profiles Reveal Global
... Our knowledge about the epigenetic changes, which accompany the determination of definitive somatic cell lineages in the mammalian embryo, are derived largely from in vitro differenti- ...
... Our knowledge about the epigenetic changes, which accompany the determination of definitive somatic cell lineages in the mammalian embryo, are derived largely from in vitro differenti- ...
Gene Section XPC (xeroderma pigmentosum, complementation group C) Atlas of Genetics and Cytogenetics
... Repair (NER) repair capacity, but the residual repair has been shown to occur specifically in transcribed genes. It is very likely that the XPC-HR23B complex is the principal damage recognition complex i.e. essential for the recognition of DNA lesions in the genome. Binding of XPC-HR23B to a DNA les ...
... Repair (NER) repair capacity, but the residual repair has been shown to occur specifically in transcribed genes. It is very likely that the XPC-HR23B complex is the principal damage recognition complex i.e. essential for the recognition of DNA lesions in the genome. Binding of XPC-HR23B to a DNA les ...
$doc.title
... 4. The rectangle below represents an electrophoresis gel used in a DNA sequencing experiment like the one discussed in class and in your text. The relevant nucleotides are labeled with a fluorescent chem ...
... 4. The rectangle below represents an electrophoresis gel used in a DNA sequencing experiment like the one discussed in class and in your text. The relevant nucleotides are labeled with a fluorescent chem ...
Formation and Repair of Complex DNA Damage Induced
... In its most common form, the double helix is wrapped nearly two turns around a histone octamer, forming a nucleosome (Figure 1). Two copies of each of the histones H2A, H2B, H3 and H4 build up the octamer and the nucleosomes are linked together with linker DNA and associated histone H1. The nucleoso ...
... In its most common form, the double helix is wrapped nearly two turns around a histone octamer, forming a nucleosome (Figure 1). Two copies of each of the histones H2A, H2B, H3 and H4 build up the octamer and the nucleosomes are linked together with linker DNA and associated histone H1. The nucleoso ...
Interaction of fission yeast ORC with essential adenine/ thymine
... DNA replication is initiated from specific sites on chromosomes called replication origins. The interaction of an initiator protein with a specific sequence near the replication origin is required for sequential assembly of other replication factors at the replication origins on chromosomes of bacte ...
... DNA replication is initiated from specific sites on chromosomes called replication origins. The interaction of an initiator protein with a specific sequence near the replication origin is required for sequential assembly of other replication factors at the replication origins on chromosomes of bacte ...
Chapter 12: Mechanisms and Regulation of Transcription I
... 4. There are two types of regulatory transcription factors a. Activating transcription factors b. Inhibitory transcription factors 5. The goal of these regulatory transcription factors is control transcription 6. They can control transcription in one of two ways a. Recruit or block efficient RNA pol ...
... 4. There are two types of regulatory transcription factors a. Activating transcription factors b. Inhibitory transcription factors 5. The goal of these regulatory transcription factors is control transcription 6. They can control transcription in one of two ways a. Recruit or block efficient RNA pol ...
Gene silencing in mammalian cells and the spread of DNA
... level persisted even when the cells were subcloned (Turker et al., 1989). These observations can best be explained by assuming that methylation for CpG sites on a given allele can be either gained or lost. By combining the results from the above studies, I proposed a model depicting a ‘dynamic equil ...
... level persisted even when the cells were subcloned (Turker et al., 1989). These observations can best be explained by assuming that methylation for CpG sites on a given allele can be either gained or lost. By combining the results from the above studies, I proposed a model depicting a ‘dynamic equil ...
Nucleosome

A nucleosome is a basic unit of DNA packaging in eukaryotes, consisting of a segment of DNA wound in sequence around eight histone protein cores. This structure is often compared to thread wrapped around a spool.Nucleosomes form the fundamental repeating units of eukaryotic chromatin, which is used to pack the large eukaryotic genomes into the nucleus while still ensuring appropriate access to it (in mammalian cells approximately 2 m of linear DNA have to be packed into a nucleus of roughly 10 µm diameter). Nucleosomes are folded through a series of successively higher order structures to eventually form a chromosome; this both compacts DNA and creates an added layer of regulatory control, which ensures correct gene expression. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones.Nucleosomes were observed as particles in the electron microscope by Don and Ada Olins and their existence and structure (as histone octamers surrounded by approximately 200 base pairs of DNA) were proposed by Roger Kornberg. The role of the nucleosome as a general gene repressor was demonstrated by Lorch et al. in vitro and by Han and Grunstein in vivo.The nucleosome core particle consists of approximately 147 base pairs of DNA wrapped in 1.67 left-handed superhelical turns around a histone octamer consisting of 2 copies each of the core histones H2A, H2B, H3, and H4. Core particles are connected by stretches of ""linker DNA"", which can be up to about 80 bp long. Technically, a nucleosome is defined as the core particle plus one of these linker regions; however the word is often synonymous with the core particle. Genome-wide nucleosome positioning maps are now available for many model organisms including mouse liver and brain.Linker histones such as H1 and its isoforms are involved in chromatin compaction and sit at the base of the nucleosome near the DNA entry and exit binding to the linker region of the DNA. Non-condensed nucleosomes without the linker histone resemble ""beads on a string of DNA"" under an electron microscope.In contrast to most eukaryotic cells, mature sperm cells largely use protamines to package their genomic DNA, most likely to achieve an even higher packaging ratio. Histone equivalents and a simplified chromatin structure have also been found in Archea, suggesting that eukaryotes are not the only organisms that use nucleosomes.