Eukaryotic gene expression
... • Eukaryotic genes require complex systems to turn them on – Chromatin structure must be relaxed in order for RNA polymerase to gain access to DNA sequence information – Eukaryotic genes are positively regulated. They are not transcribed in the absence of active mechanisms. – The regulatory componen ...
... • Eukaryotic genes require complex systems to turn them on – Chromatin structure must be relaxed in order for RNA polymerase to gain access to DNA sequence information – Eukaryotic genes are positively regulated. They are not transcribed in the absence of active mechanisms. – The regulatory componen ...
Regulation of Gene Expression
... break it down. Repressor protein combines with the available lactose (inducer) which inactivates the repressor. This allows the cell to make the enzyme. ...
... break it down. Repressor protein combines with the available lactose (inducer) which inactivates the repressor. This allows the cell to make the enzyme. ...
Chapter 18
... Overview: Conducting the Genetic Orchestra • Prokaryotes and eukaryotes alter gene expression in response to their changing environment • In multicellular eukaryotes, gene expression regulates development and is responsible for differences in cell types ...
... Overview: Conducting the Genetic Orchestra • Prokaryotes and eukaryotes alter gene expression in response to their changing environment • In multicellular eukaryotes, gene expression regulates development and is responsible for differences in cell types ...
chapter10_all
... Key Concepts • Examples in Eukaryotes • One of the two X chromosomes is inactivated in every cell of female mammals • The Y chromosome carries a master gene that causes male traits to develop in the human fetus • Flower development is orchestrated by a set of homeotic genes ...
... Key Concepts • Examples in Eukaryotes • One of the two X chromosomes is inactivated in every cell of female mammals • The Y chromosome carries a master gene that causes male traits to develop in the human fetus • Flower development is orchestrated by a set of homeotic genes ...
Transcriptome - Nematode bioinformatics. Analysis tools and data
... grid of nodes – Neighboring nodes are more similar than points far away ...
... grid of nodes – Neighboring nodes are more similar than points far away ...
The Dolan DNA Learning Center at Cold Spring Harbor Laboratory
... 4. What role does the repressor (product of the lacI gene) play in control of transcription of the lac operon? It is inhibits transcription when physically bound to the regulatory region. 5. What effect does the inducer have on the lacI gene product? It has an allosteric effect on the repressor, cha ...
... 4. What role does the repressor (product of the lacI gene) play in control of transcription of the lac operon? It is inhibits transcription when physically bound to the regulatory region. 5. What effect does the inducer have on the lacI gene product? It has an allosteric effect on the repressor, cha ...
Transcription
... • TFIIB binds adjacent to TBP and TATA box • TFIIF consists of two subunits. The larger subunit has an ATP-dependent DNA helicase activity and the small one contacts the core polymerase. • TFIIE and TFIIH are required for promoter clearance to allow RNA polymerase to commence movement away from the ...
... • TFIIB binds adjacent to TBP and TATA box • TFIIF consists of two subunits. The larger subunit has an ATP-dependent DNA helicase activity and the small one contacts the core polymerase. • TFIIE and TFIIH are required for promoter clearance to allow RNA polymerase to commence movement away from the ...
Eukaryotic Gene Control
... 2. The expression of specific genes can be inhibited by the presence of a repressor. (Negative control: repressor protein present which prevents transcription, inducer (usually a small molecule) is needed to allow initiation of transcription) 3. Inducers and repressors are small molecules that inter ...
... 2. The expression of specific genes can be inhibited by the presence of a repressor. (Negative control: repressor protein present which prevents transcription, inducer (usually a small molecule) is needed to allow initiation of transcription) 3. Inducers and repressors are small molecules that inter ...
RNA polymerase I
... glucose first. So long as glucose is present, operons such as lactose are not transcribed efficiently. Only after exhausting the supply of glucose does the bacterium fully turn on expression of the lac operon. Glucose exerts its effect, in part, by decreasing synthesis of cAMP which is required for ...
... glucose first. So long as glucose is present, operons such as lactose are not transcribed efficiently. Only after exhausting the supply of glucose does the bacterium fully turn on expression of the lac operon. Glucose exerts its effect, in part, by decreasing synthesis of cAMP which is required for ...
Chapter 10 Version #2 - Jamestown School District
... Explain how RNA is made during transcription Interpret the genetic code to determine the amino acid coded for by the codon CCU Compare the roles of the three different types of RNA during translation What is the maximum number of amino acids that could be coded for by a section of mRNA with the ...
... Explain how RNA is made during transcription Interpret the genetic code to determine the amino acid coded for by the codon CCU Compare the roles of the three different types of RNA during translation What is the maximum number of amino acids that could be coded for by a section of mRNA with the ...
HGD Gene Expression
... Nuclear export of RNA is regulated by the Cap Binding Complex (CBC) which binds exclusively to capped RNA. The CBC is then recognized by the nuclear pore complex and exported. 2. Prevention of degradation by exonucleases. Degradation of the mRNA by 5' exonucleases is prevented by functionally lookin ...
... Nuclear export of RNA is regulated by the Cap Binding Complex (CBC) which binds exclusively to capped RNA. The CBC is then recognized by the nuclear pore complex and exported. 2. Prevention of degradation by exonucleases. Degradation of the mRNA by 5' exonucleases is prevented by functionally lookin ...
Genetic Transcription & Translation Lecture PowerPoint
... Most of the RNA in cells is associated with structures known as ribosomes, the protein factories of the cells. It is the site of translation where genetic information brought by mRNA is translated into actual proteins. ...
... Most of the RNA in cells is associated with structures known as ribosomes, the protein factories of the cells. It is the site of translation where genetic information brought by mRNA is translated into actual proteins. ...
Transcription
... The structure of a bacterial RNA polymerase. Two depictions of the three-dimensional structure of a bacterial RNA polymerase, with the DNA and RNA modeled in. This RNA polymerase is formed from four different subunits, indicated by different colors (right). The DNA strand used as a template is red, ...
... The structure of a bacterial RNA polymerase. Two depictions of the three-dimensional structure of a bacterial RNA polymerase, with the DNA and RNA modeled in. This RNA polymerase is formed from four different subunits, indicated by different colors (right). The DNA strand used as a template is red, ...
Chromatin Remodeling and Gene Expression
... decrease in histone–DNA interactions. (C) Addition of ABA triggers the assembly of the ABA signaling cascade components (2) that interact with the ABRE within the phas promoter (3), leading to the recruitment of RNA Pol II and GTFs (4). New histone code modifications (H3-K4 trimethylation, H3-K14 an ...
... decrease in histone–DNA interactions. (C) Addition of ABA triggers the assembly of the ABA signaling cascade components (2) that interact with the ABRE within the phas promoter (3), leading to the recruitment of RNA Pol II and GTFs (4). New histone code modifications (H3-K4 trimethylation, H3-K14 an ...
Accounting for all the factors
... tion, stress responses and cell-cycle control, receptor pathways that drive differentiation. converge on the activation of specific families Sangamo develops zinc finger DNA-binding “You don’t have to know of transcription factors (TFs). Thus, the activity protein transcription factors (ZFP) that re ...
... tion, stress responses and cell-cycle control, receptor pathways that drive differentiation. converge on the activation of specific families Sangamo develops zinc finger DNA-binding “You don’t have to know of transcription factors (TFs). Thus, the activity protein transcription factors (ZFP) that re ...
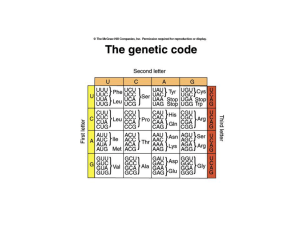
Features of the genetic code
... • Splicesome is needed to identify and catalyze the sequence of events leading to removal of the intron and rejoining of the two successive exons. The splicesome consists of snRNP (snRNA 100300 nucleotides long + proteins). Each splicesome is composed of four snRNPs together and each snRNP is five s ...
... • Splicesome is needed to identify and catalyze the sequence of events leading to removal of the intron and rejoining of the two successive exons. The splicesome consists of snRNP (snRNA 100300 nucleotides long + proteins). Each splicesome is composed of four snRNPs together and each snRNP is five s ...
Fundamentals of Cell Biology
... proteins in the cytosol and interior of these organelles help maintain these proteins in an unfolded and folded state, respectively. – Some mRNAs can be localized to specific regions of the cytosol, thereby controlling where the resulting proteins are concentrated. The actin and microtubule cytoskel ...
... proteins in the cytosol and interior of these organelles help maintain these proteins in an unfolded and folded state, respectively. – Some mRNAs can be localized to specific regions of the cytosol, thereby controlling where the resulting proteins are concentrated. The actin and microtubule cytoskel ...
PPT - Department of Computer Science
... • Particle swarm optimization (PSO) is a population based stochastic optimization technique and it is inspired by social behavior of bird flocking or fish schooling. • PSO shares many similarities with evolutionary computation techniques such as Genetic Algorithms (GA). But it is simpler and faster ...
... • Particle swarm optimization (PSO) is a population based stochastic optimization technique and it is inspired by social behavior of bird flocking or fish schooling. • PSO shares many similarities with evolutionary computation techniques such as Genetic Algorithms (GA). But it is simpler and faster ...
The Arabinose Operon
... Operons are groups of genes that function to produce proteins needed by the cell. There are two different kinds of genes in operons: Structural genes code for proteins needed for the normal operation of the cell. For example, they may be proteins needed for the breakdown of sugars. The structural ge ...
... Operons are groups of genes that function to produce proteins needed by the cell. There are two different kinds of genes in operons: Structural genes code for proteins needed for the normal operation of the cell. For example, they may be proteins needed for the breakdown of sugars. The structural ge ...
The Central Dogma of Molecular Biology
... The process of removing the intron is called splicing The intron is looped out and cut away from the exons by snRNPs (small nuclear ribonucleoprotein) (snurps) The exons are spliced together to produce the translatable mRNA The mRNA is now ready to leave the nucleus and be translated into protein ...
... The process of removing the intron is called splicing The intron is looped out and cut away from the exons by snRNPs (small nuclear ribonucleoprotein) (snurps) The exons are spliced together to produce the translatable mRNA The mRNA is now ready to leave the nucleus and be translated into protein ...
Terminator
... Types of Mutations • Point mutations (substitutions) – change in a single nucleotide – Due to redundancy of the genetic code it may change the amino acid, it may not “wobble” – Silent mutations do not change the protein Normal hemoglobin DNA ...
... Types of Mutations • Point mutations (substitutions) – change in a single nucleotide – Due to redundancy of the genetic code it may change the amino acid, it may not “wobble” – Silent mutations do not change the protein Normal hemoglobin DNA ...
chapter 19 the organization and control of eukaryotic genomes
... The amino acid (N-terminus) of each histone protein (the histone tail) extends outward from the nucleosome. ...
... The amino acid (N-terminus) of each histone protein (the histone tail) extends outward from the nucleosome. ...
PROTEIN SYNTHESIS What is a gene?
... Types of Mutations • Point mutations (substitutions) – change in a single nucleotide – Due to redundancy of the genetic code it may change the amino acid, it may not “wobble” – Silent mutations do not change the protein Normal hemoglobin DNA ...
... Types of Mutations • Point mutations (substitutions) – change in a single nucleotide – Due to redundancy of the genetic code it may change the amino acid, it may not “wobble” – Silent mutations do not change the protein Normal hemoglobin DNA ...
“Adventures in Eukaryotic Gene Expression: Transcription, Splicing, Polyadenylation, and RNAi”
... Afternoon Alumni Poster Session, 3-6 p.m. ...
... Afternoon Alumni Poster Session, 3-6 p.m. ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.