Forensic DNA Fingerprinting Kit - Bio-Rad
... 13. How important is restriction buffer concentration when doing a restriction digest? 14. Are enzymes as effective after exposure to ultraviolet (UV) light? 15. Can I mutate DNA using UV light? Does this change restriction sites? ...
... 13. How important is restriction buffer concentration when doing a restriction digest? 14. Are enzymes as effective after exposure to ultraviolet (UV) light? 15. Can I mutate DNA using UV light? Does this change restriction sites? ...
19. IMG-ER Curation Environment
... EC number and PUBMED ID – see explanation Notes are free text (goes to “note” in GenBank submission) Gene symbol is “gene name” – 4 letter abbreviation; goes to “gene” in GenBank submission ...
... EC number and PUBMED ID – see explanation Notes are free text (goes to “note” in GenBank submission) Gene symbol is “gene name” – 4 letter abbreviation; goes to “gene” in GenBank submission ...
Chapter 20
... this is used to make single stranded DNA molecules from the mRNA. mRNA gets enzymatically degraded and DNA polymerase then synthesizes a second strand of the DNA. The cDNA is modified with REs to ease the transition into plasmids and then bacterial cells. ...
... this is used to make single stranded DNA molecules from the mRNA. mRNA gets enzymatically degraded and DNA polymerase then synthesizes a second strand of the DNA. The cDNA is modified with REs to ease the transition into plasmids and then bacterial cells. ...
DETERMINATION OF NUCLEOTIDE SEQUENCES IN DNA
... comparison of a human DNA with a bovine protein, the possibility that the differences were due to some species variation, although unlikely, could not be completely excluded. For a conclusive determination of the mitochondrial code it was necessary to compare the DNA sequence of a gene with the amin ...
... comparison of a human DNA with a bovine protein, the possibility that the differences were due to some species variation, although unlikely, could not be completely excluded. For a conclusive determination of the mitochondrial code it was necessary to compare the DNA sequence of a gene with the amin ...
Teacher quality grant
... nucleus of virtually every cell. Eukaryotic cell Nucleus CHROMOSOME One or more unique pieces of DNA—circular in prokaryotes, linear in eukaryotes—that together make up an organism's genome. Chromosomes vary in length and can consist of hundreds of millions of base pairs. Humans have 23 unique chrom ...
... nucleus of virtually every cell. Eukaryotic cell Nucleus CHROMOSOME One or more unique pieces of DNA—circular in prokaryotes, linear in eukaryotes—that together make up an organism's genome. Chromosomes vary in length and can consist of hundreds of millions of base pairs. Humans have 23 unique chrom ...
DNA - Gulf Coast State College
... nucleus of virtually every cell. Eukaryotic cell Nucleus CHROMOSOME One or more unique pieces of DNA—circular in prokaryotes, linear in eukaryotes—that together make up an organism's genome. Chromosomes vary in length and can consist of hundreds of millions of base pairs. Humans have 23 unique chrom ...
... nucleus of virtually every cell. Eukaryotic cell Nucleus CHROMOSOME One or more unique pieces of DNA—circular in prokaryotes, linear in eukaryotes—that together make up an organism's genome. Chromosomes vary in length and can consist of hundreds of millions of base pairs. Humans have 23 unique chrom ...
mRNA Codon
... Proteins are vital to living organisms. They are involved in chemical reactions, oxygen transport, muscle contraction, sensory perception, blood clotting, and many other activities. The great variety of roles requires equal variety in the structure of protein molecules. This variety is achieved by m ...
... Proteins are vital to living organisms. They are involved in chemical reactions, oxygen transport, muscle contraction, sensory perception, blood clotting, and many other activities. The great variety of roles requires equal variety in the structure of protein molecules. This variety is achieved by m ...
UNITS 3 and 4 - BaysideFastTrackBiology2015
... DNA is a double-stranded molecule. The strands are connected by complementary nucleotide pairs (A-T and C-G) like rungs on a ladder. The ladder twists to form a double helix. In order for cells to make proteins, the DNA code must be transcribed (copied) to messenger RNA (mRNA). The mRNA carries the ...
... DNA is a double-stranded molecule. The strands are connected by complementary nucleotide pairs (A-T and C-G) like rungs on a ladder. The ladder twists to form a double helix. In order for cells to make proteins, the DNA code must be transcribed (copied) to messenger RNA (mRNA). The mRNA carries the ...
2) Chromatin = uncoiled DNA
... 12) _________________________is the process through which mRNA is decoded and forms a protein. 13) _________________________ is the process through which DNA transfers the code to mRNA. 14) ___________________________________ is the sugar in RNA. 15) _______________________________________ is the su ...
... 12) _________________________is the process through which mRNA is decoded and forms a protein. 13) _________________________ is the process through which DNA transfers the code to mRNA. 14) ___________________________________ is the sugar in RNA. 15) _______________________________________ is the su ...
PowerPoint-Präsentation
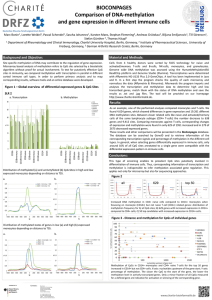
... found 4.624 genes, which showed differences in gene expression and 19.261 different DNA methylation sites. Between closer related cells like naive and activated/memory cells of the same lymphocyte subtype (CD4+ T-cells) the number decrease to 638 genes and 9.412 sites. Comparing monocytes against T- ...
... found 4.624 genes, which showed differences in gene expression and 19.261 different DNA methylation sites. Between closer related cells like naive and activated/memory cells of the same lymphocyte subtype (CD4+ T-cells) the number decrease to 638 genes and 9.412 sites. Comparing monocytes against T- ...
Ei dian otsikkoa
... Insertion site: the 5’ end of the insert shows homology with LTR sequences of the Z. mays alpha Zein gene cluster. No homology between LTR sequences and the 3’ end: rearrangement of the integration site. (Hernandez et al. (2003) Transgenic Res. 12: 179-189; Holck et al. (2002) Eur. Food Res. Tech. 2 ...
... Insertion site: the 5’ end of the insert shows homology with LTR sequences of the Z. mays alpha Zein gene cluster. No homology between LTR sequences and the 3’ end: rearrangement of the integration site. (Hernandez et al. (2003) Transgenic Res. 12: 179-189; Holck et al. (2002) Eur. Food Res. Tech. 2 ...
+ – DNA
... • Why is each person’s DNA pattern different? – sections of “junk” DNA • doesn’t code for proteins • made up of repeated patterns ...
... • Why is each person’s DNA pattern different? – sections of “junk” DNA • doesn’t code for proteins • made up of repeated patterns ...
21st 2014 Célia Miguel
... •Small RNAs contribute to posttranscriptional gene silencing by affecting mRNA stability or translation •Small RNAs contribute to transcriptional gene silencing through epigenetic modifications to chromatin ...
... •Small RNAs contribute to posttranscriptional gene silencing by affecting mRNA stability or translation •Small RNAs contribute to transcriptional gene silencing through epigenetic modifications to chromatin ...
Evolution: Mutation
... The mutation which occurs during the removal of chromosomal DNA is called a deletion of genes. An inversion happens when a section of a chromosome rotates, but the genes are still present. A translocation occurs when a section of chromosome breaks and relocates itself to a different chromosome. A su ...
... The mutation which occurs during the removal of chromosomal DNA is called a deletion of genes. An inversion happens when a section of a chromosome rotates, but the genes are still present. A translocation occurs when a section of chromosome breaks and relocates itself to a different chromosome. A su ...
DNA and Genealogy
... a process of "mixing and matching" of paired chromosomes that takes place at cell division. One or more segments may be swapped between the two chromosomes, or occasionally a segment may replace the corresponding segment on the other chromosome. This process can also occur on palindromic segments of ...
... a process of "mixing and matching" of paired chromosomes that takes place at cell division. One or more segments may be swapped between the two chromosomes, or occasionally a segment may replace the corresponding segment on the other chromosome. This process can also occur on palindromic segments of ...
Leukaemia Section t(11;19)(q23;p13.3) MLL/ACER1 Atlas of Genetics and Cytogenetics in Oncology and Haematology
... Only one case to date, a case of congenital leukemia (Lo Nigro et al., 2002). ...
... Only one case to date, a case of congenital leukemia (Lo Nigro et al., 2002). ...
Midterm #1 Study Guide
... What is the difference between mitosis and meiosis? Where do these processes occur? What are the results from each? Proteins associated with DNA in eukaryotes are called ______. Histone–DNA units are called _______. Chromatids that are attached at the centromere are called what kind of chromatids? ...
... What is the difference between mitosis and meiosis? Where do these processes occur? What are the results from each? Proteins associated with DNA in eukaryotes are called ______. Histone–DNA units are called _______. Chromatids that are attached at the centromere are called what kind of chromatids? ...
Inherited variation at the epigenetic level: paramutation from the
... of the transcription start is required to establish methylation and then expression of the active paternal allele. When this region had been replaced by one that controls methylation and allele-specific expression of the imprinted Igf2r gene, paternal inheritance of the mutated Rasgrf1 locus allowed ...
... of the transcription start is required to establish methylation and then expression of the active paternal allele. When this region had been replaced by one that controls methylation and allele-specific expression of the imprinted Igf2r gene, paternal inheritance of the mutated Rasgrf1 locus allowed ...
Gene Expression and Development
... • The other important source of developmental information is the environment around the cell, especially signals impinging on an embryonic cell from nearby cells. • In animals, these signals include contact with cell-surface molecules on neighboring cells and the binding of growth factors secreted b ...
... • The other important source of developmental information is the environment around the cell, especially signals impinging on an embryonic cell from nearby cells. • In animals, these signals include contact with cell-surface molecules on neighboring cells and the binding of growth factors secreted b ...
Assignment 2
... a. She will develop the phenotype as she ages. b. She is a carrier, and will not develop the phenotype c. She is homozygous for the wild-type allele, and hence she will not develop the phenotype d. The genotype given is not informative enough to conclude the risk. Answer: c – will remain unaffected ...
... a. She will develop the phenotype as she ages. b. She is a carrier, and will not develop the phenotype c. She is homozygous for the wild-type allele, and hence she will not develop the phenotype d. The genotype given is not informative enough to conclude the risk. Answer: c – will remain unaffected ...
Primary transcript

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor messenger RNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and complete the transcription of DNA in the nucleus of eukaryotes. Certain factors play key roles in the activation and inhibition of transcription, where they regulate primary transcript production. Transcription produces primary transcripts that are further modified by several processes. These processes include the 5' cap, 3'-polyadenylation, and alternative splicing. In particular, alternative splicing directly contributes to the diversity of mRNA found in cells. The modifications of primary transcripts have been further studied in research seeking greater knowledge of the role and significance of these transcripts. Experimental studies based on molecular changes to primary transcripts the processes before and after transcription have led to greater understanding of diseases involving primary transcripts.