Microarrays and Cancer - URMC
... expressed (turned on) in different cells under different conditions. When a gene is expressed, the genetic instructions coded in its DNA are transcribed (copied) to make messenger RNA (mRNA) molecules; then the coded instructions in messenger RNA are translated by the ribosome to make a specific pro ...
... expressed (turned on) in different cells under different conditions. When a gene is expressed, the genetic instructions coded in its DNA are transcribed (copied) to make messenger RNA (mRNA) molecules; then the coded instructions in messenger RNA are translated by the ribosome to make a specific pro ...
DNA Microarrays (Gene Chips) and Cancer - URMC
... expressed (turned on) in different cells under different conditions. When a gene is expressed, the genetic instructions coded in its DNA are transcribed (copied) to make messenger RNA (mRNA) molecules; then the coded instructions in messenger RNA are translated by the ribosome to make a specific pro ...
... expressed (turned on) in different cells under different conditions. When a gene is expressed, the genetic instructions coded in its DNA are transcribed (copied) to make messenger RNA (mRNA) molecules; then the coded instructions in messenger RNA are translated by the ribosome to make a specific pro ...
Castric et al. (MBE 2010) - GEPV
... times within dominant alleles. The relative magnitude of these two effects remains to be determined, both theoretically and empirically. A second distinctive feature of the molecular evolution of SI genes is the role played by recombination and/or gene conversion. Indeed, whether recombination occur ...
... times within dominant alleles. The relative magnitude of these two effects remains to be determined, both theoretically and empirically. A second distinctive feature of the molecular evolution of SI genes is the role played by recombination and/or gene conversion. Indeed, whether recombination occur ...
gene amplification and genomic plasticity in prokaryotes
... These structures are thought to be formed by mechanisms entailing unequal exchanges or through circle-excision and reinsertion (see Mechanisms for Gene Amplification). Tandem duplications generally do not lead to loss of gene function (2, 93). Recombination between any pair of long repeated sequence ...
... These structures are thought to be formed by mechanisms entailing unequal exchanges or through circle-excision and reinsertion (see Mechanisms for Gene Amplification). Tandem duplications generally do not lead to loss of gene function (2, 93). Recombination between any pair of long repeated sequence ...
Understanding the Scurred condition in Polled Cattle
... Understanding the Scurred condition in Polled Cattle Most calves that are horned can be identified at birth or shortly after, and almost always by weaning time. Distinguishing between smooth-polled and scurred-polled is more difficult. Scurs, a rudimentary horn growth, are often not seen until cattl ...
... Understanding the Scurred condition in Polled Cattle Most calves that are horned can be identified at birth or shortly after, and almost always by weaning time. Distinguishing between smooth-polled and scurred-polled is more difficult. Scurs, a rudimentary horn growth, are often not seen until cattl ...
The Drosophila pipsqueak gene encodes a nuclear BTB
... at (883, 884) was inferred based on Southern analysis of genomic DNA and homology to consensus sequences; the precise position of this intron was determined by Zollman et al. (1994). The genomic structure that we have determined differs somewhat from that presented by Weber et al. (1995). Sequence w ...
... at (883, 884) was inferred based on Southern analysis of genomic DNA and homology to consensus sequences; the precise position of this intron was determined by Zollman et al. (1994). The genomic structure that we have determined differs somewhat from that presented by Weber et al. (1995). Sequence w ...
Estimating the ``Effective Number of Codons`
... estimation of the ‘‘effective number of codons,’’ Nc. Several attempts have been made recently to improve Wright’s estimate of Nc, but the methods that work in cases where a gene encodes a protein not containing all amino acids with degenerate codons have not been tested against each other. In this ...
... estimation of the ‘‘effective number of codons,’’ Nc. Several attempts have been made recently to improve Wright’s estimate of Nc, but the methods that work in cases where a gene encodes a protein not containing all amino acids with degenerate codons have not been tested against each other. In this ...
A phylogenomic study of DNA repair genes, proteins
... with known functions and based on inferring evolutionary events such as gene duplications that may identify groups of genes with similar functions w22x. Specific functional predictions made for repair genes are discussed in the sections on the different repair pathways. It is important to note that ...
... with known functions and based on inferring evolutionary events such as gene duplications that may identify groups of genes with similar functions w22x. Specific functional predictions made for repair genes are discussed in the sections on the different repair pathways. It is important to note that ...
ANOVA and the Bootstrap - Computational Diagnostics Group
... Fit MHo to the observed data and calculate residuals and fitted values. Create a new data set by resampling with replacement from the residuals. Add the new residuals to the fitted values. Calculate t* by fitting Model MA to the new data set Repeat both steps many times (1000 times) Bootstrap p – va ...
... Fit MHo to the observed data and calculate residuals and fitted values. Create a new data set by resampling with replacement from the residuals. Add the new residuals to the fitted values. Calculate t* by fitting Model MA to the new data set Repeat both steps many times (1000 times) Bootstrap p – va ...
Robust gene silencing mediated by antisense small RNAs in the
... nitrosative stress (32), heat shock (33), histone acetylation (34), DNA methylation (35) or tissue invasion (25,36). EHI_197520 had abundant sRNAs and low gene expression in the HM-1:IMSS strain, but no sRNAs and high gene expression in the Rahman strain. Conversely, EHI_025700 had abundant sRNAs an ...
... nitrosative stress (32), heat shock (33), histone acetylation (34), DNA methylation (35) or tissue invasion (25,36). EHI_197520 had abundant sRNAs and low gene expression in the HM-1:IMSS strain, but no sRNAs and high gene expression in the Rahman strain. Conversely, EHI_025700 had abundant sRNAs an ...
Multiple Domains Exist within the Upstream Activator
... direct the synthesis and secretion of unusual amino acid and sugar derivatives called opines. The inciting strain of Agrobacterium can utilize these opines as a carbon and sometimes a nitrogen source. Agrobacterium strains can be classified based upon their ability to catabolize particular opines. ( ...
... direct the synthesis and secretion of unusual amino acid and sugar derivatives called opines. The inciting strain of Agrobacterium can utilize these opines as a carbon and sometimes a nitrogen source. Agrobacterium strains can be classified based upon their ability to catabolize particular opines. ( ...
Green Fluorescent Protein
... GFP is used within various fields of biology like molecular biology, neuroscience and cell biology. It can be used as a reporter gene, cell marker, fusion tag, and for quantitative monitoring of gene expression. GFP’s fluorescence characteristic can also be used in methods designed for sensing vario ...
... GFP is used within various fields of biology like molecular biology, neuroscience and cell biology. It can be used as a reporter gene, cell marker, fusion tag, and for quantitative monitoring of gene expression. GFP’s fluorescence characteristic can also be used in methods designed for sensing vario ...
Lecture-Mic 623-Plasmids-Listeria - Home
... We determined the sequence and genetic organization of plasmid pIP823, which contains the dfrD gene; dfrD confers high-level trimethoprim resistance to Listeria monocytogenes BM4293 by synthesis of dihydrofolate reductase type S2. pIP823 possessed all the features of the pUB110/pC194 plasmid family ...
... We determined the sequence and genetic organization of plasmid pIP823, which contains the dfrD gene; dfrD confers high-level trimethoprim resistance to Listeria monocytogenes BM4293 by synthesis of dihydrofolate reductase type S2. pIP823 possessed all the features of the pUB110/pC194 plasmid family ...
Gene Section TYR (tyrosinase (oculocutaneous albinism IA)) Atlas of Genetics and Cytogenetics
... iris in patients with OCA1A is light blue or gray and the retina lacks pigmentation as well. Tyrosinase null patients have greatly reduced visual acuity accompanied by nystagmus, strabismus, and usually photophobia (Ray et al., 2007). Patients with OCA1B present with varying levels of pigment. The h ...
... iris in patients with OCA1A is light blue or gray and the retina lacks pigmentation as well. Tyrosinase null patients have greatly reduced visual acuity accompanied by nystagmus, strabismus, and usually photophobia (Ray et al., 2007). Patients with OCA1B present with varying levels of pigment. The h ...
A versatile toolbox for PCR-based tagging of yeast genes: new
... new selection markers have been introduced. The tags include the so far brightest and most yeast-optimized version of the red fluorescent protein, called RedStar2, as well as all other commonly used fluorescent proteins and tags used for the detection and purification of proteins and protein complex ...
... new selection markers have been introduced. The tags include the so far brightest and most yeast-optimized version of the red fluorescent protein, called RedStar2, as well as all other commonly used fluorescent proteins and tags used for the detection and purification of proteins and protein complex ...
TCA (Krebs) Cycle
... ©Copyright 1999-2004 by Gene C. Lavers No part of this presentation may be reproduced by any mechanical, photographic, or electronic process, or in the form of a phonographic recording, nor may it be stored in a retrieval system, transmitted, or otherwise copied for public or private use, without wr ...
... ©Copyright 1999-2004 by Gene C. Lavers No part of this presentation may be reproduced by any mechanical, photographic, or electronic process, or in the form of a phonographic recording, nor may it be stored in a retrieval system, transmitted, or otherwise copied for public or private use, without wr ...
NSPKU
... • A protein substitute contains NO Phe... But it contains all of the other amino acids in protein that your child needs. • Most protein substitutes also contain vitamins, minerals and other important nutrients to ensure your child is getting what they need. The protein substitute is an extremely imp ...
... • A protein substitute contains NO Phe... But it contains all of the other amino acids in protein that your child needs. • Most protein substitutes also contain vitamins, minerals and other important nutrients to ensure your child is getting what they need. The protein substitute is an extremely imp ...
Gene Name
... hydrocarbon (Ah) receptor is involved in the induction of several enzymes that participate in xenobiotic metabolism. The ligand-free, cytosolic form of the Ah receptor is complexed to heat shock protein 90. Binding of ligand, which includes dioxin and polycyclic aromatic hydrocarbons, results in tra ...
... hydrocarbon (Ah) receptor is involved in the induction of several enzymes that participate in xenobiotic metabolism. The ligand-free, cytosolic form of the Ah receptor is complexed to heat shock protein 90. Binding of ligand, which includes dioxin and polycyclic aromatic hydrocarbons, results in tra ...
magamtol talalt cikkek
... regulates an enormous variety of cellular functions through the interaction of its catalytic subunit (PP1c) with over fifty different established or putative regulatory subunits. Most of these target PP1c to specific subcellular locations and interact with a small hydrophobic groove on the surface o ...
... regulates an enormous variety of cellular functions through the interaction of its catalytic subunit (PP1c) with over fifty different established or putative regulatory subunits. Most of these target PP1c to specific subcellular locations and interact with a small hydrophobic groove on the surface o ...
Gene Section LMO2 (LIM domain only 2 (rhombotin-like 1))
... but could act as a bridging molecule bringing together different DNA binding factors (TAL1, LDB1, E12/E47, GATA1) that are essential for hematopoiesis (e.g. in the erythroid complex). This interaction is critical for the regulation of red blood cell development in early stages of hematopoiesis. TAL1 ...
... but could act as a bridging molecule bringing together different DNA binding factors (TAL1, LDB1, E12/E47, GATA1) that are essential for hematopoiesis (e.g. in the erythroid complex). This interaction is critical for the regulation of red blood cell development in early stages of hematopoiesis. TAL1 ...
Genes - Gerstein Lab Publications
... retropseudogenes and found that genes with a high number of retropseudogene copies tend to be widely expressed, highly conserved and low in (G+C) content. With the complete genomes of more than 30 prokaryotes and 4 eukaryotes (including the Caenorhabditis elegans genome [19]) now published, we have ...
... retropseudogenes and found that genes with a high number of retropseudogene copies tend to be widely expressed, highly conserved and low in (G+C) content. With the complete genomes of more than 30 prokaryotes and 4 eukaryotes (including the Caenorhabditis elegans genome [19]) now published, we have ...
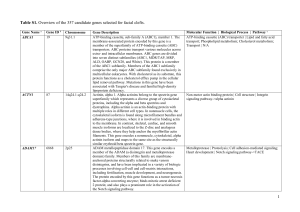
(1977) New Genes for Resistance to the Brown Planthopper in Rice
... with TN1 were crossed with IRl154-243, a selection with the recessive gene, bph 2 for resistance (7). Cultivars, whose F~ hybrids with TN1 were r~sistant, were crossed with IR1539-823, a dwarf selection that is homozygous for the dominant gene, Bph 1 for resistance (5). To determine the allelic rela ...
... with TN1 were crossed with IRl154-243, a selection with the recessive gene, bph 2 for resistance (7). Cultivars, whose F~ hybrids with TN1 were r~sistant, were crossed with IR1539-823, a dwarf selection that is homozygous for the dominant gene, Bph 1 for resistance (5). To determine the allelic rela ...