3.1.1.4 Proteins
... muscle proteins that work together to cause a muscle to contract. There are proteins in cell membranes that help identify a cell or serve as a receptor. Adrenalin and insulin are two examples of hormones that are made of protein. All proteins have a special shape that is the result of the interactio ...
... muscle proteins that work together to cause a muscle to contract. There are proteins in cell membranes that help identify a cell or serve as a receptor. Adrenalin and insulin are two examples of hormones that are made of protein. All proteins have a special shape that is the result of the interactio ...
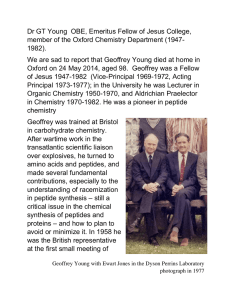
DrGTYoungOBE
... After wartime work in the transatlantic scientific liaison over explosives, he turned to amino acids and peptides, and made several fundamental contributions, especially to the understanding of racemization in peptide synthesis – still a critical issue in the chemical synthesis of peptides and prote ...
... After wartime work in the transatlantic scientific liaison over explosives, he turned to amino acids and peptides, and made several fundamental contributions, especially to the understanding of racemization in peptide synthesis – still a critical issue in the chemical synthesis of peptides and prote ...
Unit 1 PPT 1 (2a Proteomics)
... composed of introns and exons. • Introns are the non-coding sequence of the mRNA and will not be expressed in the protein molecule. They are spliced out (removed) from the mRNA. • Exons are the coding sequence and will be expressed in the protein molecule. • RNA splicing in detail. ...
... composed of introns and exons. • Introns are the non-coding sequence of the mRNA and will not be expressed in the protein molecule. They are spliced out (removed) from the mRNA. • Exons are the coding sequence and will be expressed in the protein molecule. • RNA splicing in detail. ...
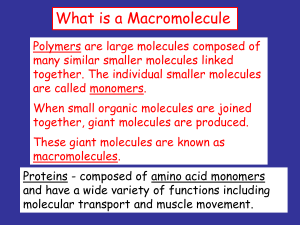
What is a Macromolecule
... enzymes - are proteins that facilitate biochemical reactions. They are often referred to as catalysts because they speed up chemical reactions. ...
... enzymes - are proteins that facilitate biochemical reactions. They are often referred to as catalysts because they speed up chemical reactions. ...
proteomics - Sigma
... of a potentially exposed, immunogenic internal sequence for antibody generation. Surface regions or regions of high accessibility often border helical or extended secondary structure regions. In addition, sequence regions with b-turn or amphipathic helix character have been found to be antigenic. Pe ...
... of a potentially exposed, immunogenic internal sequence for antibody generation. Surface regions or regions of high accessibility often border helical or extended secondary structure regions. In addition, sequence regions with b-turn or amphipathic helix character have been found to be antigenic. Pe ...
14.5 Uncommon Amino Acids
... make up a chain of protein • Different sequences of peptide and protein molecules allows for the protein to carry out its functions • The formula for calculating the possible numbers of peptides and proteins for a chain of n amino acids by raising it to the 20th power • 20ⁿ ...
... make up a chain of protein • Different sequences of peptide and protein molecules allows for the protein to carry out its functions • The formula for calculating the possible numbers of peptides and proteins for a chain of n amino acids by raising it to the 20th power • 20ⁿ ...
An insight into the (un)stable protein formulation
... time. Often, it starts in with the conformational change of the protein and is typically accompanied by the formation of an antiparallel β-sheet structure. These conformational changes can be detected very sensitively using FT-IR spectroscopy. So, this technique enables the identification of unstabl ...
... time. Often, it starts in with the conformational change of the protein and is typically accompanied by the formation of an antiparallel β-sheet structure. These conformational changes can be detected very sensitively using FT-IR spectroscopy. So, this technique enables the identification of unstabl ...
Supplementary Methods
... The raw data files were converted to the Mascot generic format and searched with the Mascot search engine (http://www.matrixscience.com) against the IPI human protein database (http://www.ebi.ac.uk). Carbamidomethylation was selected as a fixed modification. Oxidation of methionine, N-acetylation of ...
... The raw data files were converted to the Mascot generic format and searched with the Mascot search engine (http://www.matrixscience.com) against the IPI human protein database (http://www.ebi.ac.uk). Carbamidomethylation was selected as a fixed modification. Oxidation of methionine, N-acetylation of ...
A Novel Scoring Function for Predicting the Conformation of Pairs of
... proteins are tightly packed. We present a scoring function and a computational methodology for predicting the tertiary fold of a pair of α-helices, such that its chances of being tightly packed are maximized. Since the number of TM protein structures solved to date is small, it seems unlikely that a ...
... proteins are tightly packed. We present a scoring function and a computational methodology for predicting the tertiary fold of a pair of α-helices, such that its chances of being tightly packed are maximized. Since the number of TM protein structures solved to date is small, it seems unlikely that a ...
Repetitive Patterns in Proteins
... known repeats (REP, Mocca, TPRpred) - De novo detection of internal sequence repeats by comparing the protein sequence to itself (HHepID) ...
... known repeats (REP, Mocca, TPRpred) - De novo detection of internal sequence repeats by comparing the protein sequence to itself (HHepID) ...
Document
... These pathways may lead to other genes being switched on or off Mass Spectrometry is key to probing the proteome ...
... These pathways may lead to other genes being switched on or off Mass Spectrometry is key to probing the proteome ...
LOYOLA COLLEGE (AUTONOMOUS), CHENNAI – 600 034
... 7. Operons are not found in prokaryotes. 8. Genes are upregulated in cancers. 9. Nitrosylation is very important for cell cycle progression. 10. Metabolomics and metagenomics are same. III Complete the following ...
... 7. Operons are not found in prokaryotes. 8. Genes are upregulated in cancers. 9. Nitrosylation is very important for cell cycle progression. 10. Metabolomics and metagenomics are same. III Complete the following ...
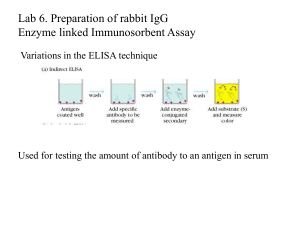
Enzyme-linked secondary antibodies
... Range of separation of proteins depends on the percentage of polyacrylamide used (typical range 12-16%). ...
... Range of separation of proteins depends on the percentage of polyacrylamide used (typical range 12-16%). ...
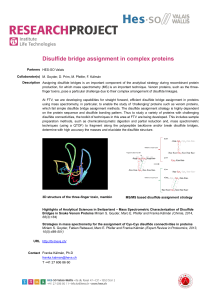
Disulfide bridge assignment in complex proteins - HES
... using mass spectrometry, in particular, to enable the study of 'challenging' proteins such as venom proteins, which fail simple disulfide bridge assignment methods. The disulfide assignment strategy is highly dependent on the protein sequence and disulfide bonding pattern. Thus to study a variety of ...
... using mass spectrometry, in particular, to enable the study of 'challenging' proteins such as venom proteins, which fail simple disulfide bridge assignment methods. The disulfide assignment strategy is highly dependent on the protein sequence and disulfide bonding pattern. Thus to study a variety of ...
Amino Acids Placemat
... of proteins. There are 20 amino acids — each with a different shape and chemical property. As they join together in a distinct sequence — specified by your DNA — they spontaneously fold into a compact shape following basic principles of chemistry and physics. ...
... of proteins. There are 20 amino acids — each with a different shape and chemical property. As they join together in a distinct sequence — specified by your DNA — they spontaneously fold into a compact shape following basic principles of chemistry and physics. ...
elastin - MBBS Students Club
... has the same amino acid sequence as the normal protein; that is, their primary structures are identical but its secondary structure is dominated by beta conformation is insoluble in all but the strongest solvents is highly resistant to digestion by proteases ...
... has the same amino acid sequence as the normal protein; that is, their primary structures are identical but its secondary structure is dominated by beta conformation is insoluble in all but the strongest solvents is highly resistant to digestion by proteases ...
ELASTIN - Rihs.com.pk
... has the same amino acid sequence as the normal protein; that is, their primary structures are identical but its secondary structure is dominated by beta conformation is insoluble in all but the strongest solvents is highly resistant to digestion by proteases ...
... has the same amino acid sequence as the normal protein; that is, their primary structures are identical but its secondary structure is dominated by beta conformation is insoluble in all but the strongest solvents is highly resistant to digestion by proteases ...
View video content as a PDF
... The Final 3-Dimensional Shape of the Protein Once the secondary structures of a protein have been folded, the model must be given the correct overall shape. When doing this it is very useful to refer back to the online visualization environment. This display can be edited to match what the final phy ...
... The Final 3-Dimensional Shape of the Protein Once the secondary structures of a protein have been folded, the model must be given the correct overall shape. When doing this it is very useful to refer back to the online visualization environment. This display can be edited to match what the final phy ...
Protein Structure
... Not all proteins folded into stable structures Intrinsically Disordered Proteins (IDPs) have regions favoring disorder IDP regions tend to lack hydrophobic residues Rich in polar amino acids and proline IDPs may favor adaptation to binding another protein IDPs may favor being modified IDPs may be mo ...
... Not all proteins folded into stable structures Intrinsically Disordered Proteins (IDPs) have regions favoring disorder IDP regions tend to lack hydrophobic residues Rich in polar amino acids and proline IDPs may favor adaptation to binding another protein IDPs may favor being modified IDPs may be mo ...
MALDI Target Spotting for Proteomics Research
... Fast and automated protein characterization is a key issue in proteomics research for drug target discovery. Large numbers of individual proteins are separated by two-dimensional gel chromatography to obtain individual protein spots. Often, the resulting protein spots are then picked, digested and a ...
... Fast and automated protein characterization is a key issue in proteomics research for drug target discovery. Large numbers of individual proteins are separated by two-dimensional gel chromatography to obtain individual protein spots. Often, the resulting protein spots are then picked, digested and a ...
simulating protein analysis using gel electrophoresis
... A technique known as gel electrophoresis is widely used to analyze the size of macromolecules. These size differences can be used for evolutionary analysis as well as the analysis of a number of other critical questions regarding both proteins and DNA. Gel electrophoresis works on two relatively sim ...
... A technique known as gel electrophoresis is widely used to analyze the size of macromolecules. These size differences can be used for evolutionary analysis as well as the analysis of a number of other critical questions regarding both proteins and DNA. Gel electrophoresis works on two relatively sim ...
Applications of spectroscopy
... Why Laser T-jump? • The introduction of pulsed lasers excitation as triggers of the biochemical processes brought dramatic improvement in the experimental time resolution. However, this methodology is inapplicable to molecules without suitable ...
... Why Laser T-jump? • The introduction of pulsed lasers excitation as triggers of the biochemical processes brought dramatic improvement in the experimental time resolution. However, this methodology is inapplicable to molecules without suitable ...
Paper background for Students
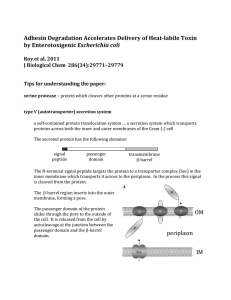
... The resulting fusion protein contains three domains: a. EtpA b. 10 amino acids of the myc protein sequence (a protein “tag) c. 6 histidine residues (a protein “tag”) This is useful because the protein can be purified using immobilized antibodies against the myc tag or the polyhistidine tag. Alternat ...
... The resulting fusion protein contains three domains: a. EtpA b. 10 amino acids of the myc protein sequence (a protein “tag) c. 6 histidine residues (a protein “tag”) This is useful because the protein can be purified using immobilized antibodies against the myc tag or the polyhistidine tag. Alternat ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.