Translation - e
... The sedimentation coefficient s of a particle is used to characterize its behaviour in sedimentation processes, notably centrifugation. It is defined as the ratio of a particle's sedimentation velocity to the acceleration that is applied to it. The sedimentation speed υt (in ms−1) is also known as t ...
... The sedimentation coefficient s of a particle is used to characterize its behaviour in sedimentation processes, notably centrifugation. It is defined as the ratio of a particle's sedimentation velocity to the acceleration that is applied to it. The sedimentation speed υt (in ms−1) is also known as t ...
RNA-Seq analysis
... mRNAs, non-coding RNAs and small RNAs Determine the transcriptional structure of genes in terms of: Start sites 5′ and 3′ ends Splicing patterns Other post-transcriptional modifications Quantification of expression levels and comparison ...
... mRNAs, non-coding RNAs and small RNAs Determine the transcriptional structure of genes in terms of: Start sites 5′ and 3′ ends Splicing patterns Other post-transcriptional modifications Quantification of expression levels and comparison ...
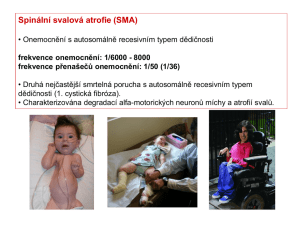
Snímek 1
... genes of the affected snRNPs are and how this specifically affects motor neuron function (indicated by a question mark (?)). One possibility is that the critical target gene is specific to motor neuron system. Alternatively, a function of critical importance to motor neurons could be disrupted. In a ...
... genes of the affected snRNPs are and how this specifically affects motor neuron function (indicated by a question mark (?)). One possibility is that the critical target gene is specific to motor neuron system. Alternatively, a function of critical importance to motor neurons could be disrupted. In a ...
Identification of functional domains in Arabidopsis thaliana mRNA
... has been proposed that the availability of the 7-methyl guanosine cap in mRNA is a dominant feature in competition for limited translation initiation factors, primarily eIF4E (1). Cap-independent translation mechanisms such as re-initiation and internal initiation generally lack the efficiency associa ...
... has been proposed that the availability of the 7-methyl guanosine cap in mRNA is a dominant feature in competition for limited translation initiation factors, primarily eIF4E (1). Cap-independent translation mechanisms such as re-initiation and internal initiation generally lack the efficiency associa ...
Translation
... endomembrane system and proteins that are secreted from the cell • Ribosomes are identical and can switch from free to bound Copyright © 2008 Pearson Education Inc., publishing as Pearson Benjamin Cummings ...
... endomembrane system and proteins that are secreted from the cell • Ribosomes are identical and can switch from free to bound Copyright © 2008 Pearson Education Inc., publishing as Pearson Benjamin Cummings ...
The RNA world meets behavior: AfiI pre
... mutation? One intrinsic advantage of editing a nucleotide over having change ‘hard-wired’ into the genome through mutation is the regulation of the degree to which a coding position is modified within messages. Certain pre-mRNA editing sites vary greatly in the frequency with which their editing is ...
... mutation? One intrinsic advantage of editing a nucleotide over having change ‘hard-wired’ into the genome through mutation is the regulation of the degree to which a coding position is modified within messages. Certain pre-mRNA editing sites vary greatly in the frequency with which their editing is ...
Protein Synthesis_MJH
... termination of polypeptide synthesis. It must be noted that genetic code is degenerate that is each amino acid is usually specified by more than one codon. Eukaryotic mRNA almost always contain a single ORF. In contrast, prokaryotic mRNA frequently contain two or more ORFs & hence can encode multi ...
... termination of polypeptide synthesis. It must be noted that genetic code is degenerate that is each amino acid is usually specified by more than one codon. Eukaryotic mRNA almost always contain a single ORF. In contrast, prokaryotic mRNA frequently contain two or more ORFs & hence can encode multi ...
Zinc finger proteins
... In contrast, fingers 4–5–6 form an open, extended structure running along one side of the DNA. Of these, only finger 5 makes contacts with bases in the major groove. The flanking fingers, 4 and 6, appear to serve primarily as spacer elements. Odd S. Gabrielsen ...
... In contrast, fingers 4–5–6 form an open, extended structure running along one side of the DNA. Of these, only finger 5 makes contacts with bases in the major groove. The flanking fingers, 4 and 6, appear to serve primarily as spacer elements. Odd S. Gabrielsen ...
Gene Section IGF1 (Insulin-Like Growth Factor 1 (Somatomedin C))
... such distinctions have yet to be definitively identified) (Adapted from Mills et al., 2007; Philippou et al., 2007). ...
... such distinctions have yet to be definitively identified) (Adapted from Mills et al., 2007; Philippou et al., 2007). ...
9th Lecture Updated
... Its unique binding site, located on 23S ribosomal RNA of the 50S subunit, results in no cross-resistance with other drug classes Resistance is caused by mutation of the linezolid binding site on 23S ribosomal RNA, with subsequent decreased binding to the target site Pharmacokinetics Linezoli ...
... Its unique binding site, located on 23S ribosomal RNA of the 50S subunit, results in no cross-resistance with other drug classes Resistance is caused by mutation of the linezolid binding site on 23S ribosomal RNA, with subsequent decreased binding to the target site Pharmacokinetics Linezoli ...
Ribosome Profiling
... TIS. From the recent RP experiments primarily targeted to map TIS, it becomes very clear that the mechanism of translational initiation is far more complex than that was previously thought. Dramatic increase in the number of non-AUG start or initiation at 5ʹ UTR sites are documented through RP exper ...
... TIS. From the recent RP experiments primarily targeted to map TIS, it becomes very clear that the mechanism of translational initiation is far more complex than that was previously thought. Dramatic increase in the number of non-AUG start or initiation at 5ʹ UTR sites are documented through RP exper ...
RNA Interference and Small Interfering RNAs
... possible by the introduction of transgenes composed ing at a considerable distance. In plants, grafting experiof inverted repeats which produce dsRNA hairpins after tranments[48, 49] as well as the localized introduction of transgenes scription. In C. elegans, for which targeted recombination-based ...
... possible by the introduction of transgenes composed ing at a considerable distance. In plants, grafting experiof inverted repeats which produce dsRNA hairpins after tranments[48, 49] as well as the localized introduction of transgenes scription. In C. elegans, for which targeted recombination-based ...
lac
... mutated, will grow on lactose. • However they make β-galactosidase all of the time. These mutants that have lost the ability to control gene expression are called constitutive mutants. They are also recessive. ...
... mutated, will grow on lactose. • However they make β-galactosidase all of the time. These mutants that have lost the ability to control gene expression are called constitutive mutants. They are also recessive. ...
RNAP IIO
... mutated SRB proteins may abolish the effect of CTD deletions SRBs = components of the Mediator - more later ...
... mutated SRB proteins may abolish the effect of CTD deletions SRBs = components of the Mediator - more later ...
Lecture 21 trp operon Chap26
... 26.10 The Operator Competes with LowAffinity Sites to Bind Repressor • In the absence of inducer, the operator has an affinity for repressor that is 107 times that of a low-affinity site. • The level of 10 repressor tetramers per cell ensures that the operator is bound by repressor 96% of the time. ...
... 26.10 The Operator Competes with LowAffinity Sites to Bind Repressor • In the absence of inducer, the operator has an affinity for repressor that is 107 times that of a low-affinity site. • The level of 10 repressor tetramers per cell ensures that the operator is bound by repressor 96% of the time. ...
PDF
... that initiated buds always developed the normal tubular appearance characteristic of later stages. This supports the earlier suggestion (Clarkson & Wolpert, 1967) that bud elongation must be interpreted in terms of tissue movement rather than growth. In addition, tentacles were frequently observed o ...
... that initiated buds always developed the normal tubular appearance characteristic of later stages. This supports the earlier suggestion (Clarkson & Wolpert, 1967) that bud elongation must be interpreted in terms of tissue movement rather than growth. In addition, tentacles were frequently observed o ...
Whole-transcriptome RNAseq analysis from minute amount of total
... it is a challenge to acquire sufficient amounts of highquality tissue specimens for genomic characterization of tumors (4). In early development studies on mouse embryos, there is insufficient RNA to analyze the transcriptome of the very low number of primordial germ cells (PGCs). It is also challengi ...
... it is a challenge to acquire sufficient amounts of highquality tissue specimens for genomic characterization of tumors (4). In early development studies on mouse embryos, there is insufficient RNA to analyze the transcriptome of the very low number of primordial germ cells (PGCs). It is also challengi ...
Sequence Alignment - Bilkent University
... all reporter mRNAs in a cell free translation system, Alu RNP acts as a general inhibitor of protein translation due to conformational changes in each. SRP mediates a transient delay in translation by blocking the elongation step, Alu RNP inhibits translation by reducing initiation. ...
... all reporter mRNAs in a cell free translation system, Alu RNP acts as a general inhibitor of protein translation due to conformational changes in each. SRP mediates a transient delay in translation by blocking the elongation step, Alu RNP inhibits translation by reducing initiation. ...
Nuclear–chloroplast
... genes which influence chloroplast development. These mutants include those with defects in differentiation and greening (dag), chloroplast and leaf development (dcl) and chlorophyll accumulation (pale cress, pac). Each of these mutants suggests that the nucleus can sense the physiological status of ...
... genes which influence chloroplast development. These mutants include those with defects in differentiation and greening (dag), chloroplast and leaf development (dcl) and chlorophyll accumulation (pale cress, pac). Each of these mutants suggests that the nucleus can sense the physiological status of ...
Flowers - Oregon State University
... •The RNA silencing signal is amplified through a combination of target cleavage, siRNA primed polymerization, and subsequent cleavage by DCL ...
... •The RNA silencing signal is amplified through a combination of target cleavage, siRNA primed polymerization, and subsequent cleavage by DCL ...
Эволюция генетического кода
... The UG doublet does not change its conformation in the contemporary genetic code and it seems that it was not prone to do this before. There is no evidence of uracil preferably being in the enol form, but, at the same time, nothing points to a greater stability of the keto form in the absence of ade ...
... The UG doublet does not change its conformation in the contemporary genetic code and it seems that it was not prone to do this before. There is no evidence of uracil preferably being in the enol form, but, at the same time, nothing points to a greater stability of the keto form in the absence of ade ...
View document as PDF
... Teaching Points When proteins fold into their tertiary structures, there are often subdivisions within the protein, designated as domains, which are characterized by similar features or motifs. One such motif is the zinc finger, in which a specific domain of the protein is arranged into a “finger-li ...
... Teaching Points When proteins fold into their tertiary structures, there are often subdivisions within the protein, designated as domains, which are characterized by similar features or motifs. One such motif is the zinc finger, in which a specific domain of the protein is arranged into a “finger-li ...
Teaching Notes
... 6. The teacher will assess each group based upon their presentation. 7. The teacher should review the concepts with their students: a. The tRNA and mRNA interact via complementary base pairing b. The directionality of reading the RNA strand is 5’ to 3’, while that for proteins/peptides is N- to C. c ...
... 6. The teacher will assess each group based upon their presentation. 7. The teacher should review the concepts with their students: a. The tRNA and mRNA interact via complementary base pairing b. The directionality of reading the RNA strand is 5’ to 3’, while that for proteins/peptides is N- to C. c ...