- ITA Heidelberg
... currently knows more than 20 000 di erent proteins in man (HPRD 2005). They have many functions: structure proteins, enzymes, hormones, transport proteins, protection proteins, tractiles, toxines, and so on. The sequence of amino acids determines the biological function of proteins. Proteins consist ...
... currently knows more than 20 000 di erent proteins in man (HPRD 2005). They have many functions: structure proteins, enzymes, hormones, transport proteins, protection proteins, tractiles, toxines, and so on. The sequence of amino acids determines the biological function of proteins. Proteins consist ...
„Biochemical reconstitution of protein complexes involved in
... Until today over 20 different proteins were identified to be important in this process, but the very core of FeS cluster assembly complex is formed by molecular scaffold protein Isu1, cysteine desulfurase Nfs1(Isd11) and frataxin Yfh1. Isu1 serves as a place of cluster biosynthesis, Nfs1(Isd11) is a ...
... Until today over 20 different proteins were identified to be important in this process, but the very core of FeS cluster assembly complex is formed by molecular scaffold protein Isu1, cysteine desulfurase Nfs1(Isd11) and frataxin Yfh1. Isu1 serves as a place of cluster biosynthesis, Nfs1(Isd11) is a ...
Introduction to Studying Proteins
... The actual protein product is produced during translation. Ribosomes use mRNA and tRNA molecules to assemble the a.a’s in the correct sequence. ...
... The actual protein product is produced during translation. Ribosomes use mRNA and tRNA molecules to assemble the a.a’s in the correct sequence. ...
We propose a frequent pattern-based algorithm for predicting
... Abstract: We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub ...
... Abstract: We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub ...
Chapter 5: Biological Molecules Molecules of Life • All life made up
... Carboxyl group Amino group Side Chain (R group) – accounts for different properties Structure & Function o Functional protein consists of 1 or more polypeptides coiled, twisted, & folded into a unique shape o Amino acid order determines protein’s 3-D structure, which determines function 4 ...
... Carboxyl group Amino group Side Chain (R group) – accounts for different properties Structure & Function o Functional protein consists of 1 or more polypeptides coiled, twisted, & folded into a unique shape o Amino acid order determines protein’s 3-D structure, which determines function 4 ...
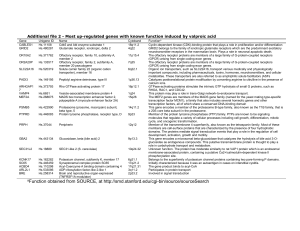
Table - BioMed Central
... transcription factor MCM1), a family that also includes several homeotic genes and other transcription factors, all of which share a conserved DNA-binding domain This gene encodes a member of the proteasome B-type family, also known as the T1B family, that is a 20S core beta subunit in the proteasom ...
... transcription factor MCM1), a family that also includes several homeotic genes and other transcription factors, all of which share a conserved DNA-binding domain This gene encodes a member of the proteasome B-type family, also known as the T1B family, that is a 20S core beta subunit in the proteasom ...
The Biology of
... helix and beta sheets • (A) is example of an alpha helix. The hydrogen bonds (dotted lines) are between oxygen atoms (red) and hydrogen atoms (white) (shown in this case as occurring every fourth pair of amino acids along the protein). • (B) shows examples of beta-sheets held together by hydrogen bo ...
... helix and beta sheets • (A) is example of an alpha helix. The hydrogen bonds (dotted lines) are between oxygen atoms (red) and hydrogen atoms (white) (shown in this case as occurring every fourth pair of amino acids along the protein). • (B) shows examples of beta-sheets held together by hydrogen bo ...
Protein Synthesis
... Proteins are chains of amino acids 20 different amino acids Combinations of amino acids determine a proteins purpose ...
... Proteins are chains of amino acids 20 different amino acids Combinations of amino acids determine a proteins purpose ...
Auxiliary proteins of photosystem II: tuning the enzyme for optimal
... The core of Photosystem II (PS II) is made up of two reaction center proteins, D1 (PsbA) and D2 (PsbD) and two chlorophyll a-binding antenna proteins, CP47 (PsbB) and CP43 (PsbC). These proteins have homologues in anoxygenic photosynthetic bacterial reaction centers; however, PS II has an increased ...
... The core of Photosystem II (PS II) is made up of two reaction center proteins, D1 (PsbA) and D2 (PsbD) and two chlorophyll a-binding antenna proteins, CP47 (PsbB) and CP43 (PsbC). These proteins have homologues in anoxygenic photosynthetic bacterial reaction centers; however, PS II has an increased ...
“Building” proteins!!
... The multi-coloured beads you have will be your amino acids and the much smaller single-coloured beads will be the bond joining the amino acids. You also have strings of different strength to use for different models. Among your materials you will find additional model making materials and tools such ...
... The multi-coloured beads you have will be your amino acids and the much smaller single-coloured beads will be the bond joining the amino acids. You also have strings of different strength to use for different models. Among your materials you will find additional model making materials and tools such ...
Biosynthesis and degradation of proteins
... Protease inhibitors • IAPs are proteins that block apoptosis by binding to and inhibiting caspases. The apoptosis-stimulating protein Smac antagonizes the effect of IAPs on caspases. • TIMPs are inhibitors of metalloproteases that are secreted by cells. A domain of the inhibitor protein interacts w ...
... Protease inhibitors • IAPs are proteins that block apoptosis by binding to and inhibiting caspases. The apoptosis-stimulating protein Smac antagonizes the effect of IAPs on caspases. • TIMPs are inhibitors of metalloproteases that are secreted by cells. A domain of the inhibitor protein interacts w ...
Chapter 3 USU - BEHS Science
... Its not just chemical formula, it’s the shape of the molecule that lets it do its “job”. ...
... Its not just chemical formula, it’s the shape of the molecule that lets it do its “job”. ...
AP Biology
... 11. Describe the structure of an amino acid. What is the alpha carbon? Do any of the amino acids contain alpha carbons that are not asymmetric? 12. What is an R group? Discuss the various properties that the R group (side chains) possess. Give an example of each type of amino acid. 13. Describe the ...
... 11. Describe the structure of an amino acid. What is the alpha carbon? Do any of the amino acids contain alpha carbons that are not asymmetric? 12. What is an R group? Discuss the various properties that the R group (side chains) possess. Give an example of each type of amino acid. 13. Describe the ...
Abstract The cytoskeleton is a cellular structure comprised of three
... Abstract The cytoskeleton is a cellular structure comprised of three types of protein filaments called microfilaments, intermediate filaments and microtubules respectively. These filaments are highly dynamic and can change their organisation and properties according to the current needs of a cell. T ...
... Abstract The cytoskeleton is a cellular structure comprised of three types of protein filaments called microfilaments, intermediate filaments and microtubules respectively. These filaments are highly dynamic and can change their organisation and properties according to the current needs of a cell. T ...
02/13
... such as DNA and RNA polymerases. They are recognized by nuclear pore proteins for transport into nucleus. ...
... such as DNA and RNA polymerases. They are recognized by nuclear pore proteins for transport into nucleus. ...
www.rcsd.k12.ca.us
... carbon atoms surrounded by as many hydrogen atoms as possible (bad guys – solid at room temp) Unsaturated fats have double carbon bonds instead of so many bonds with hydrogen (good guys) Hydrogenated fats have had hydrogen added to unsaturated fats. ...
... carbon atoms surrounded by as many hydrogen atoms as possible (bad guys – solid at room temp) Unsaturated fats have double carbon bonds instead of so many bonds with hydrogen (good guys) Hydrogenated fats have had hydrogen added to unsaturated fats. ...
Cartoon modeling of proteins
... formation + motor protein separation + control sequences) Self assembly of viruses from their coat proteins ...
... formation + motor protein separation + control sequences) Self assembly of viruses from their coat proteins ...
Cartoon modeling of proteins
... formation + motor protein separation + control sequences) Self assembly of viruses from their coat proteins ...
... formation + motor protein separation + control sequences) Self assembly of viruses from their coat proteins ...
amino acids
... • different for each amino acid • confers unique chemical properties to each amino acid ...
... • different for each amino acid • confers unique chemical properties to each amino acid ...
Notes Guide Part 2
... Protein- Amino acids join by _________________________________ rxn to form dipeptides and polypeptides. ...
... Protein- Amino acids join by _________________________________ rxn to form dipeptides and polypeptides. ...
Proteins as Supramolecular Building Blocks
... st ructures and active nanoscaffolds. Two model systems are being explored: a TIM barrel enzyme, representing the most common protein fold and therefore scaffold for activity; and the peroxiredoxins, a family of proteins that have already revealed themselves to have unique self-assembly properties c ...
... st ructures and active nanoscaffolds. Two model systems are being explored: a TIM barrel enzyme, representing the most common protein fold and therefore scaffold for activity; and the peroxiredoxins, a family of proteins that have already revealed themselves to have unique self-assembly properties c ...
Biological Molecules
... chains of small molecules. In proteins, these small molecules are not identical. protein molecule ...
... chains of small molecules. In proteins, these small molecules are not identical. protein molecule ...
Protein

Proteins (/ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within living organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific three-dimensional structure that determines its activity.A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than about 20-30 residues, are rarely considered to be proteins and are commonly called peptides, or sometimes oligopeptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in a protein is defined by the sequence of a gene, which is encoded in the genetic code. In general, the genetic code specifies 20 standard amino acids; however, in certain organisms the genetic code can include selenocysteine and—in certain archaea—pyrrolysine. Shortly after or even during synthesis, the residues in a protein are often chemically modified by posttranslational modification, which alters the physical and chemical properties, folding, stability, activity, and ultimately, the function of the proteins. Sometimes proteins have non-peptide groups attached, which can be called prosthetic groups or cofactors. Proteins can also work together to achieve a particular function, and they often associate to form stable protein complexes.Once formed, proteins only exist for a certain period of time and are then degraded and recycled by the cell's machinery through the process of protein turnover. A protein's lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells. Abnormal and or misfolded proteins are degraded more rapidly either due to being targeted for destruction or due to being unstable.Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of organisms and participate in virtually every process within cells. Many proteins are enzymes that catalyze biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses, cell adhesion, and the cell cycle. Proteins are also necessary in animals' diets, since animals cannot synthesize all the amino acids they need and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that are then used in metabolism.Proteins may be purified from other cellular components using a variety of techniques such as ultracentrifugation, precipitation, electrophoresis, and chromatography; the advent of genetic engineering has made possible a number of methods to facilitate purification. Methods commonly used to study protein structure and function include immunohistochemistry, site-directed mutagenesis, X-ray crystallography, nuclear magnetic resonance and mass spectrometry.