Sequence variation: Looking for effects of
... Drosophila [2]; it is not known whether this is true for the fluctuating fitness model. These considerations raise two questions: Are such patterns general? And how can we test between different possible explanations of the patterns? Surveys of DNA sequence variation are rapidly being extended beyon ...
... Drosophila [2]; it is not known whether this is true for the fluctuating fitness model. These considerations raise two questions: Are such patterns general? And how can we test between different possible explanations of the patterns? Surveys of DNA sequence variation are rapidly being extended beyon ...
Serine Protease Mechanism
... • In the absence of an enzyme, potential reactant molecules adopt a NAC only about 0.0001% of the time • On the other hand, NACs have been shown to form in enzyme active sites from 1% to 70% of the time Figure 14.7 NACs are characterized as having reacting atoms within 3.2 Å and an approach angle o ...
... • In the absence of an enzyme, potential reactant molecules adopt a NAC only about 0.0001% of the time • On the other hand, NACs have been shown to form in enzyme active sites from 1% to 70% of the time Figure 14.7 NACs are characterized as having reacting atoms within 3.2 Å and an approach angle o ...
Contextual Genetic Algorithms: Evolving Developmental Rules
... The discovery of messenger RNA (mRNA) molecules containing information not coded in DNA, first persuaded researchers in molecular biology that some mechanism in the cell might be responsible for posttranscriptional alteration of genetic information; this mechanism was called 'RNA Editing' [2, 1986]. ...
... The discovery of messenger RNA (mRNA) molecules containing information not coded in DNA, first persuaded researchers in molecular biology that some mechanism in the cell might be responsible for posttranscriptional alteration of genetic information; this mechanism was called 'RNA Editing' [2, 1986]. ...
BIO 110 Survey of Biology QZM 3 Q 150701abbr.2
... c. Proteins d. Lipids e. Salt 3. To what does the term "polypeptide" specifically refer? a. organic molecules linked by dehydration synthesis b. organic monomers covalently bonded c. amino acids linked by hydrolysis d. carbohydrates with a hydrogen bond holding them together e. none of the above Gen ...
... c. Proteins d. Lipids e. Salt 3. To what does the term "polypeptide" specifically refer? a. organic molecules linked by dehydration synthesis b. organic monomers covalently bonded c. amino acids linked by hydrolysis d. carbohydrates with a hydrogen bond holding them together e. none of the above Gen ...
Analyzing Evolvability To Anticipate New Pathogens
... opposite. Thus, it is advantageous for individual cells within a population to express slightly different antigens along their surfaces, enabling at least some of them to escape when they are all exposed to a host immune surveillance system. Therefore, VlsE is said to have a high potential for evolv ...
... opposite. Thus, it is advantageous for individual cells within a population to express slightly different antigens along their surfaces, enabling at least some of them to escape when they are all exposed to a host immune surveillance system. Therefore, VlsE is said to have a high potential for evolv ...
Analysis of the DNA microarray hybridization images using
... paper aims at detecting the spots on the image of DNA microarray. A critical stage of analysis of the DNA array hybridization pictures is determination positions of the spots. Typical hybridization pictures contain, besides desirable objects (spots), some amount of noise. Considering requirement of ...
... paper aims at detecting the spots on the image of DNA microarray. A critical stage of analysis of the DNA array hybridization pictures is determination positions of the spots. Typical hybridization pictures contain, besides desirable objects (spots), some amount of noise. Considering requirement of ...
Molecular Biology of the Cell
... providing the initial means to identify and number each chromosome. ...
... providing the initial means to identify and number each chromosome. ...
DUAL TRAFFICKING PATHWAYS OF CONNEXINS TO GAP …
... gene pairs, osaB is independently transcribed ...
... gene pairs, osaB is independently transcribed ...
Consortium for Educational Communication
... altering the equilibrium between the reactants and the products. Although, the rate at which the chemical equilibrium is established is enhanced. The enzymes affect only the rate and not the direction. They can accelerate the reaction in either direction. Many enzymes which are required for the synt ...
... altering the equilibrium between the reactants and the products. Although, the rate at which the chemical equilibrium is established is enhanced. The enzymes affect only the rate and not the direction. They can accelerate the reaction in either direction. Many enzymes which are required for the synt ...
13-2 PowerPoint
... The Molecular Basis of Heredity One of the most interesting discoveries of molecular biology is the near-universal nature of the genetic code. Although some organisms show slight variations in the amino acids assigned to particular codons, the code is always read three bases at a time and in the sam ...
... The Molecular Basis of Heredity One of the most interesting discoveries of molecular biology is the near-universal nature of the genetic code. Although some organisms show slight variations in the amino acids assigned to particular codons, the code is always read three bases at a time and in the sam ...
Gene7-02
... Exon is any segment of an interrupted gene that is represented in the mature RNA product. Intron is a segment of DNA that is transcribed, but removed from within the transcript by splicing together the sequences (exons) on either side of it. RNA splicing is the process of excising the sequences in R ...
... Exon is any segment of an interrupted gene that is represented in the mature RNA product. Intron is a segment of DNA that is transcribed, but removed from within the transcript by splicing together the sequences (exons) on either side of it. RNA splicing is the process of excising the sequences in R ...
The Drosophila Ribosomal Protein S6 Gene Includes a 3
... can result in proteins with repeated functional domains. In work presented elsewhere (Stewart and Denell 1993), we have shown that two transposon-induced mutations causing a loss of growth control of the Drosophila larval hematopoietic organs affect the gene encoding ribosomal protein S6 (rpS6 ) . T ...
... can result in proteins with repeated functional domains. In work presented elsewhere (Stewart and Denell 1993), we have shown that two transposon-induced mutations causing a loss of growth control of the Drosophila larval hematopoietic organs affect the gene encoding ribosomal protein S6 (rpS6 ) . T ...
chapter 20 lecture (ppt file)
... Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. ...
... Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. ...
pdf file
... • All organisms use the same 20 aa • Each codon specifies a particular aa Figure 10.8A Copyright © 2003 Pearson Education, Inc. publishing as Benjamin Cummings ...
... • All organisms use the same 20 aa • Each codon specifies a particular aa Figure 10.8A Copyright © 2003 Pearson Education, Inc. publishing as Benjamin Cummings ...
Protein Sequence Databases
... as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for ...
... as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for ...
Gene Section PCSK5 (proprotein convertase subtilisin/kexin type 5) Atlas of Genetics and Cytogenetics
... Other names: PC5, PC6, PC6A, SPC6 HGNC (Hugo): PCSK5 Location: 9q21.13 ...
... Other names: PC5, PC6, PC6A, SPC6 HGNC (Hugo): PCSK5 Location: 9q21.13 ...
DNA-Directed Antibody Immobilization for
... piezoelectric arrayer. All antibodies were spotted in PBS with 50 mM Trehalose. All ssDNA surface probes were spotted at a concentration of 30 μM in sodium phosphate buffer (150 mM, pH = 8.5). The spotted chips were kept overnight in the spotter chamber at 67% humidity. Following the overnight immobi ...
... piezoelectric arrayer. All antibodies were spotted in PBS with 50 mM Trehalose. All ssDNA surface probes were spotted at a concentration of 30 μM in sodium phosphate buffer (150 mM, pH = 8.5). The spotted chips were kept overnight in the spotter chamber at 67% humidity. Following the overnight immobi ...
The nucleotide sequence of the tnpA gene completes the sequence
... known genes and major open reading frames. The terminal inverted repeats and the major promoter (39) are marked. The gene designated merP is that described as merC in (26), and has been renamed because the merC gene originally identified in plasmid R100 by genetic criteria (40) corresponds to a read ...
... known genes and major open reading frames. The terminal inverted repeats and the major promoter (39) are marked. The gene designated merP is that described as merC in (26), and has been renamed because the merC gene originally identified in plasmid R100 by genetic criteria (40) corresponds to a read ...
Mutations in the parkin gene cause autosomal
... (lodged under accession number AB009973 in the DDBJ database). The parkin gene consists of 12 exons, with exon 7 corresponding to the putative exon J-17 obtained by the initial exon trapping. Based on these data, we designed 14 sets of PCR primers to amplify each of 12 exons and determined the size ...
... (lodged under accession number AB009973 in the DDBJ database). The parkin gene consists of 12 exons, with exon 7 corresponding to the putative exon J-17 obtained by the initial exon trapping. Based on these data, we designed 14 sets of PCR primers to amplify each of 12 exons and determined the size ...
Poly-ε-caprolactone electrospun nanofiber mesh as a
... and high mortality [2]. It is still an urgent need to develop new therapeutic strategies for this deadly malignancy. Gene therapy, emerging as a promising treatment strategy for many acquired or inherited diseases including cancers, is a milestone of bio-medical field [3,4]. Gene delivery methodolog ...
... and high mortality [2]. It is still an urgent need to develop new therapeutic strategies for this deadly malignancy. Gene therapy, emerging as a promising treatment strategy for many acquired or inherited diseases including cancers, is a milestone of bio-medical field [3,4]. Gene delivery methodolog ...
AQA(B) AS Module 2 - heckgrammar.co.uk
... • The two strands are joined together by hydrogen bonds between the bases. The bases therefore form base pairs, which are like rungs of a ladder. • The base pairs are specific. A only binds to T (and T with A), and C only binds to G (and G with C). These are called complementary base pairs (or somet ...
... • The two strands are joined together by hydrogen bonds between the bases. The bases therefore form base pairs, which are like rungs of a ladder. • The base pairs are specific. A only binds to T (and T with A), and C only binds to G (and G with C). These are called complementary base pairs (or somet ...
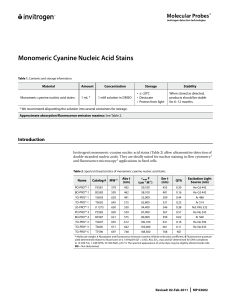
Monomeric Cyanine Nucleic Acid Stains
... The fluorescence spectra of the monomeric cyanine nucleic acid stains cover the entire visible wavelength range, as shown in Figure 1 and summarized in Table 2. The table also lists optical filter sets and laser line sources suitable for excitation and detection of these dyes. These dyes may also be ...
... The fluorescence spectra of the monomeric cyanine nucleic acid stains cover the entire visible wavelength range, as shown in Figure 1 and summarized in Table 2. The table also lists optical filter sets and laser line sources suitable for excitation and detection of these dyes. These dyes may also be ...
Deoxyribozyme
_DNAzyme.png?width=300)
Deoxyribozymes, also called DNA enzymes, DNAzymes, or catalytic DNA, are DNA oligonucleotides that are capable of catalyzing specific chemical reactions, similar to the action of other biological enzymes, such as proteins or ribozymes (enzymes composed of RNA).However, in contrast to the abundance of protein enzymes in biological systems and the discovery of biological ribozymes in the 1980s,there are no known naturally occurring deoxyribozymes.Deoxyribozymes should not be confused with DNA aptamers which are oligonucleotides that selectively bind a target ligand, but do not catalyze a subsequent chemical reaction.With the exception of ribozymes, nucleic acid molecules within cells primarily serve as storage of genetic information due to its ability to form complementary base pairs, which allows for high-fidelity copying and transfer of genetic information. In contrast, nucleic acid molecules are more limited in their catalytic ability, in comparison to protein enzymes, to just three types of interactions: hydrogen bonding, pi stacking, and metal-ion coordination. This is due to the limited number of functional groups of the nucleic acid monomers: while proteins are built from up to twenty different amino acids with various functional groups, nucleic acids are built from just four chemically similar nucleobases. In addition, DNA lacks the 2'-hydroxyl group found in RNA which limits the catalytic competency of deoxyribozymes even in comparison to ribozymes.In addition to the inherent inferiority of DNA catalytic activity, the apparent lack of naturally occurring deoxyribozymes may also be due to the primarily double-stranded conformation of DNA in biological systems which would limit its physical flexibility and ability to form tertiary structures, and so would drastically limit the ability of double-stranded DNA to act as a catalyst; though there are a few known instances of biological single-stranded DNA such as multicopy single-stranded DNA (msDNA), certain viral genomes, and the replication fork formed during DNA replication. Further structural differences between DNA and RNA may also play a role in the lack of biological deoxyribozymes, such as the additional methyl group of the DNA base thymidine compared to the RNA base uracil or the tendency of DNA to adopt the B-form helix while RNA tends to adopt the A-form helix. However, it has also been shown that DNA can form structures that RNA cannot, which suggests that, though there are differences in structures that each can form, neither is inherently more or less catalytic due to their possible structural motifs.