Type III Secretion System
... The effector genes are not linked between species showing that they are independent of the genes for TTSS protein secretion This allows the bacteria to adapt to host countermeasures or to a new host This is an important process because each different type of bacteria has a preferential niche, ...
... The effector genes are not linked between species showing that they are independent of the genes for TTSS protein secretion This allows the bacteria to adapt to host countermeasures or to a new host This is an important process because each different type of bacteria has a preferential niche, ...
CS689-domains - faculty.cs.tamu.edu
... no hydrophobic core glucagon (30 res, a-helix); dis-ordered in solution unraveling, conformational sampling NMR studies of peptides? 10-aa SCF recognition peptide disorder of p53 fragment in soln by NMR on the contrary, 17-residue fragment from N-terminal domain of ubiquitin folds into beta-hairpin ...
... no hydrophobic core glucagon (30 res, a-helix); dis-ordered in solution unraveling, conformational sampling NMR studies of peptides? 10-aa SCF recognition peptide disorder of p53 fragment in soln by NMR on the contrary, 17-residue fragment from N-terminal domain of ubiquitin folds into beta-hairpin ...
ProSyn
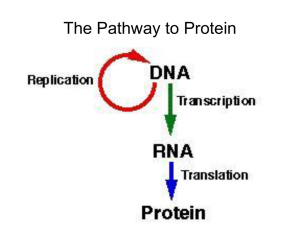
... – Uracil instead of thymine – Ribose instead of deoxyribose – Shorter than DNA – Folds to form some double-stranded regions (A-U, C-G) ...
... – Uracil instead of thymine – Ribose instead of deoxyribose – Shorter than DNA – Folds to form some double-stranded regions (A-U, C-G) ...
Transcription
... 2) 3’ tail: poly-A tail (adenine); protection; recognition; transport 3) RNA splicing: exons (expressed sequences) kept,introns (intervening sequences) spliced out; – snRNPs (small nuclear ribonucleoproteins) – join to form spliceosomes – recognize splice sites – release introns/join exons ...
... 2) 3’ tail: poly-A tail (adenine); protection; recognition; transport 3) RNA splicing: exons (expressed sequences) kept,introns (intervening sequences) spliced out; – snRNPs (small nuclear ribonucleoproteins) – join to form spliceosomes – recognize splice sites – release introns/join exons ...
Modification of Genes and Proteins - sharonap-cellrepro-p2
... Protein factors identify damage DNA is unwound Faulty area is cut out and the bases are removed DNA is synthesized to match that of the ...
... Protein factors identify damage DNA is unwound Faulty area is cut out and the bases are removed DNA is synthesized to match that of the ...
Bioinformatics and Functional Genomics, Chapter 8, Part 1
... Inferred by curator Inferred from direct assay Inferred from electronic annotation Inferred from expression pattern Inferred from genetic interaction Inferred from mutant phenotype Inferred from physical interaction Inferred from sequence or structural similarity Non-traceable author statement No bi ...
... Inferred by curator Inferred from direct assay Inferred from electronic annotation Inferred from expression pattern Inferred from genetic interaction Inferred from mutant phenotype Inferred from physical interaction Inferred from sequence or structural similarity Non-traceable author statement No bi ...
105 Quantitative Analysis of Crude Protein
... This analysis method is applied for determination of crude proteins in products containing proteins, such as food products. ...
... This analysis method is applied for determination of crude proteins in products containing proteins, such as food products. ...
Anti-CD30 human IL-2 fusion proteins display strong and specific
... Although therapy of CD30-positive lymphomas such as classical Hodgkin lymphoma and anaplastic large cell lymphoma has been improved considerably during the last decades, patients suffer from high toxicity of current therapeutic regimens. Since CD30 expression is very restricted, CD30-positive tumors ...
... Although therapy of CD30-positive lymphomas such as classical Hodgkin lymphoma and anaplastic large cell lymphoma has been improved considerably during the last decades, patients suffer from high toxicity of current therapeutic regimens. Since CD30 expression is very restricted, CD30-positive tumors ...
Job - Cloudfront.net
... 7) Which organelle converts sunlight into sugar? 8) Which organelle creates ribosomes? 9) Which organelle fuses with the cell membrane to release proteins? 10)Which molecule holds the information to make a protein? ...
... 7) Which organelle converts sunlight into sugar? 8) Which organelle creates ribosomes? 9) Which organelle fuses with the cell membrane to release proteins? 10)Which molecule holds the information to make a protein? ...
Figure 5.1 Rapid Diffusion of Membrane Proteins The fluid mosaic
... The fluid mosaic model of cell membranes, described by Singer and Nicolson (1972), was critical to understanding biological membranes as proteins floating in a phospholipid matrix. Integral to this model was earlier work by Frye and Edidin (1970). These researchers examined the movement of proteins ...
... The fluid mosaic model of cell membranes, described by Singer and Nicolson (1972), was critical to understanding biological membranes as proteins floating in a phospholipid matrix. Integral to this model was earlier work by Frye and Edidin (1970). These researchers examined the movement of proteins ...
Oct - CSIR-NEIST, Jorhat
... surface of the virus, and is covered with carbohydrate chains that hide it from the immune system. It is also a highly dynamic protein that snaps into a different shape when it binds to a cell surface, dragging the virus and cell close enough to each other that the membranes fuse. The structure show ...
... surface of the virus, and is covered with carbohydrate chains that hide it from the immune system. It is also a highly dynamic protein that snaps into a different shape when it binds to a cell surface, dragging the virus and cell close enough to each other that the membranes fuse. The structure show ...
The Cell Membrane
... -it controls “selects” what can enter or leave the cell -some materials are allowed through others aren’t ...
... -it controls “selects” what can enter or leave the cell -some materials are allowed through others aren’t ...
doc CHEE_370_HW_1_
... proteins from Escherichia coli (which has very low levels of K+ in its cytoplasm). Which amino acids do you think are enriched in Halobacterium proteins and why? (Hint: which amino acids could best neutralize the positive charges due to K+?) 5. (10 points) RNA and DNA are similar types of macromolec ...
... proteins from Escherichia coli (which has very low levels of K+ in its cytoplasm). Which amino acids do you think are enriched in Halobacterium proteins and why? (Hint: which amino acids could best neutralize the positive charges due to K+?) 5. (10 points) RNA and DNA are similar types of macromolec ...
1. The term peptidyltransferase relates to A. base additions during
... elongation factors binding to the large ribosomal subunit. ...
... elongation factors binding to the large ribosomal subunit. ...
Unit 03 Macromolecule Review
... 6. How are monosaccharides important to plants? To humans? 7. How is cellulose important to plants? To humans? 8. How is starch important to plants? To humans? 9. What organisms use glycogen? For what? 10. A person has dangerously low blood-glucose and needs glucose quickly. You have the choice of g ...
... 6. How are monosaccharides important to plants? To humans? 7. How is cellulose important to plants? To humans? 8. How is starch important to plants? To humans? 9. What organisms use glycogen? For what? 10. A person has dangerously low blood-glucose and needs glucose quickly. You have the choice of g ...
Eukaryotic Gene Regulation
... 1. For each of the following, is the statement true for prokaryotes or eukaryotes? a. Each gene is regulated separately by its own promoter b. DNA is not packed very extensively c. Regulatory proteins are made in the nucleus but act in the cytoplasm d. Less complex promoter structure with less regul ...
... 1. For each of the following, is the statement true for prokaryotes or eukaryotes? a. Each gene is regulated separately by its own promoter b. DNA is not packed very extensively c. Regulatory proteins are made in the nucleus but act in the cytoplasm d. Less complex promoter structure with less regul ...
Expression system
... • Often a rare protease cut site is added to the fusion partner • Eg., For small peptides poly-arginine,Histidine tail ...
... • Often a rare protease cut site is added to the fusion partner • Eg., For small peptides poly-arginine,Histidine tail ...
walk the dogma notes - Nutley Public Schools
... - Function- Carries genetic info from the DNA in the nucleus to the cytoplasm 2) Transfer RNA (tRNA) - Structure: Hairpin Loop - Function: Binds and carries specific amino acids 3) Ribosomal RNA (rRNA) - Structure: Globular - Function: Combines with proteins to form ribosomes ...
... - Function- Carries genetic info from the DNA in the nucleus to the cytoplasm 2) Transfer RNA (tRNA) - Structure: Hairpin Loop - Function: Binds and carries specific amino acids 3) Ribosomal RNA (rRNA) - Structure: Globular - Function: Combines with proteins to form ribosomes ...
From Gene to Protein
... Free ribosomes: synthesize proteins that stay in cytosol and function there Bound ribosomes (to ER): make proteins of endomembrane system (nuclear envelope, ER, Golgi, lysosomes, vacuoles, plasma membrane) & proteins for ...
... Free ribosomes: synthesize proteins that stay in cytosol and function there Bound ribosomes (to ER): make proteins of endomembrane system (nuclear envelope, ER, Golgi, lysosomes, vacuoles, plasma membrane) & proteins for ...
Recitation 3 - MIT OpenCourseWare
... patterns in different region of polypeptide chains and is predominantly stabilized by hydrogen bonds. The different interactions between the side chain groups of the amino acids determine the 3dimensional tertiary structure of proteins. Quaternary structure results when two or more polypeptide chain ...
... patterns in different region of polypeptide chains and is predominantly stabilized by hydrogen bonds. The different interactions between the side chain groups of the amino acids determine the 3dimensional tertiary structure of proteins. Quaternary structure results when two or more polypeptide chain ...
Chapter 5: Biological Molecules Molecules of Life • All life made up
... o Each has unique amino acid sequence; can be a few to more than a thousand Amino Acid Structure o -Carbon bonded to: Hydrogen Carboxyl group Amino group Side Chain (R group) – accounts for different properties Structure & Function o Functional protein consists of 1 or more polypeptides ...
... o Each has unique amino acid sequence; can be a few to more than a thousand Amino Acid Structure o -Carbon bonded to: Hydrogen Carboxyl group Amino group Side Chain (R group) – accounts for different properties Structure & Function o Functional protein consists of 1 or more polypeptides ...
5.3 Presentation: Protein Synthesis
... • Cells respond to their environments by producing different types and amounts of proteins • The cell produces proteins that are structural (forms part of cell materials) or functional (enzymes and hormones). • All of an organisms cells have the same DNA, but the cells differ on the expression of th ...
... • Cells respond to their environments by producing different types and amounts of proteins • The cell produces proteins that are structural (forms part of cell materials) or functional (enzymes and hormones). • All of an organisms cells have the same DNA, but the cells differ on the expression of th ...
Lecture Slides for Protein Structure
... • Each domain folds by mechanisms similar to those above. ...
... • Each domain folds by mechanisms similar to those above. ...
SR protein

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are ""S"" and ""R"" respectively. SR proteins are 50-300 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS binding domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.SR proteins were discovered in the 1990s in Drosophila and in amphibian oocytes, and later in humans. In general, metazoans appear to have SR proteins and unicellular organisms lack SR proteins.SR proteins are important in constitutive and alternative pre-mRNA splicing, mRNA export, genome stabilization, nonsense-mediated decay, and translation. SR proteins alternatively splice pre-mRNA by preferentially selecting different splice sites on the pre-mRNA strands to create multiple mRNA transcripts from one pre-mRNA transcript. Once splicing is complete the SR protein may or may not remain attached to help shuttle the mRNA strand out of the nucleus. As RNA Polymerase II is transcribing DNA into RNA, SR proteins attach to newly made pre-mRNA to prevent the pre-mRNA from binding to the coding DNA strand to increase genome stabilization. Topoisomerase I and SR proteins also interact to increase genome stabilization. SR proteins can control the concentrations of specific mRNA that is successfully translated into protein by selecting for nonsense-mediated decay codons during alternative splicing. SR proteins can alternatively splice NMD codons into its own mRNA transcript to auto-regulate the concentration of SR proteins. Through the mTOR pathway and interactions with polyribosomes, SR proteins can increase translation of mRNA.Ataxia telangiectasia, neurofibromatosis type 1, several cancers, HIV-1, and spinal muscular atrophy have all been linked to alternative splicing by SR proteins.