Gene to protein
... TRANSLATION = RNA → PROTEINS (Occurs on RIBOSOMES in CYTOPLASM in both PROKARYOTES & EUKARYOTES) Specific AMINO ACYL tRNA SYNTHETASES added amino acids to correct tRNA’s TRANSLATION & TRANSCRIPTION happen simultaneously in PROKARYOTES ...
... TRANSLATION = RNA → PROTEINS (Occurs on RIBOSOMES in CYTOPLASM in both PROKARYOTES & EUKARYOTES) Specific AMINO ACYL tRNA SYNTHETASES added amino acids to correct tRNA’s TRANSLATION & TRANSCRIPTION happen simultaneously in PROKARYOTES ...
Ch 1617 Study Guide - Dublin City Schools
... TRANSLATION = RNA → PROTEINS (Occurs on RIBOSOMES in CYTOPLASM in both PROKARYOTES & EUKARYOTES) Specific AMINO ACYL tRNA SYNTHETASES added amino acids to correct tRNA’s TRANSLATION & TRANSCRIPTION happen simultaneously in PROKARYOTES ...
... TRANSLATION = RNA → PROTEINS (Occurs on RIBOSOMES in CYTOPLASM in both PROKARYOTES & EUKARYOTES) Specific AMINO ACYL tRNA SYNTHETASES added amino acids to correct tRNA’s TRANSLATION & TRANSCRIPTION happen simultaneously in PROKARYOTES ...
Cellular compartmentalization
... This complex diffuses until it reaches a contact site, where it is treaded through both channel proteins. Once inside the cell the protein refolds and the signal sequence is cleaved off. ...
... This complex diffuses until it reaches a contact site, where it is treaded through both channel proteins. Once inside the cell the protein refolds and the signal sequence is cleaved off. ...
Unit Three “Cell Proliferation and Genetics”
... Categories of RNA • Ribosomal RNA (rRNA) – DNA serves as template for production of rRNA; formed in the nucleus of a cell; moves into the cytoplasm to bond with proteins; rRNA + proteins make up Ribosomes (site of protein synthesis) • Messenger RNA (mRNA) – DNA serves as template for production of ...
... Categories of RNA • Ribosomal RNA (rRNA) – DNA serves as template for production of rRNA; formed in the nucleus of a cell; moves into the cytoplasm to bond with proteins; rRNA + proteins make up Ribosomes (site of protein synthesis) • Messenger RNA (mRNA) – DNA serves as template for production of ...
File - Thomas Tallis School
... round in shape such as enzymes and haemoglobin. The exact shapes of proteins can be very important in how they work. Proteins are made of 20 different kinds of amino acids joined in a chain. The chains can contain between 50 and many thousands of amino acids, but most are a few hundred amino acids i ...
... round in shape such as enzymes and haemoglobin. The exact shapes of proteins can be very important in how they work. Proteins are made of 20 different kinds of amino acids joined in a chain. The chains can contain between 50 and many thousands of amino acids, but most are a few hundred amino acids i ...
Macromolecules
... Macromolecules are formed when monomers are linked together to form longer chains called polymers. The same process of making and breaking polymers is found in all living organisms. ...
... Macromolecules are formed when monomers are linked together to form longer chains called polymers. The same process of making and breaking polymers is found in all living organisms. ...
File
... …Then the mRNA code reads like this… UAG-CAU-GCG-UUA-ACG The enzyme RNA polymerase separates two strands of a DNA double helix and builds a strand of RNA using RNA nucleotides. Remember, RNA coding: ...
... …Then the mRNA code reads like this… UAG-CAU-GCG-UUA-ACG The enzyme RNA polymerase separates two strands of a DNA double helix and builds a strand of RNA using RNA nucleotides. Remember, RNA coding: ...
Gene Expression
... The same three steps are repeated until the “stop” codon is read. 1. An amino acid is placed in position on the “A” site of the ribosome 2. The peptide bond is formed. 3. The peptide moves over to the “P” site so that the “A” site is available for the next amino acid. (The old tRNA is released.) ...
... The same three steps are repeated until the “stop” codon is read. 1. An amino acid is placed in position on the “A” site of the ribosome 2. The peptide bond is formed. 3. The peptide moves over to the “P” site so that the “A” site is available for the next amino acid. (The old tRNA is released.) ...
Proteins
... …Then the mRNA code reads like this… UAG-CAU-GCG-UUA-ACG The enzyme RNA polymerase separates two strands of a DNA double helix and builds a strand of RNA using RNA nucleotides. Remember, RNA coding: ...
... …Then the mRNA code reads like this… UAG-CAU-GCG-UUA-ACG The enzyme RNA polymerase separates two strands of a DNA double helix and builds a strand of RNA using RNA nucleotides. Remember, RNA coding: ...
Proteins – Organic/Macromolecule #3
... giving the cell and organism the ability to move, examples are actin and myosin that help muscles contract. Like the organic molecules the elements that make up proteins are carbon, hydrogen, oxygen, nitrogen and sulfur. All proteins have a special shape that is the result of the interactions among ...
... giving the cell and organism the ability to move, examples are actin and myosin that help muscles contract. Like the organic molecules the elements that make up proteins are carbon, hydrogen, oxygen, nitrogen and sulfur. All proteins have a special shape that is the result of the interactions among ...
Proteins – Organic/Macromolecule #3
... giving the cell and organism the ability to move, examples are actin and myosin that help muscles contract. Like the organic molecules the elements that make up proteins are carbon, hydrogen, oxygen, nitrogen and sulfur. All proteins have a special shape that is the result of the interactions among ...
... giving the cell and organism the ability to move, examples are actin and myosin that help muscles contract. Like the organic molecules the elements that make up proteins are carbon, hydrogen, oxygen, nitrogen and sulfur. All proteins have a special shape that is the result of the interactions among ...
Chapt. 3-Proteins - University of New England
... • Example: enzyme binding sites, substrate binding sites ...
... • Example: enzyme binding sites, substrate binding sites ...
Chapter 2APa Study Guide
... coordination & control; antibodies, clotting proteins 15. Why is cholesterol an important component of the cell membrane? allows cell memb to slide & change shape as needed 16. Be able to label/recognize the components of an AA. What is the importance of the R group? ...
... coordination & control; antibodies, clotting proteins 15. Why is cholesterol an important component of the cell membrane? allows cell memb to slide & change shape as needed 16. Be able to label/recognize the components of an AA. What is the importance of the R group? ...
The macromolecular sites of action through which drugs
... proteins may be obtained by first considering what forces drive protein folding since these 2 processes share many common characteristics. The observed tertiary or quaternary structure of proteins is in large part a consequence of the hydrophobic effect. This means that proteins tend to bury hydroph ...
... proteins may be obtained by first considering what forces drive protein folding since these 2 processes share many common characteristics. The observed tertiary or quaternary structure of proteins is in large part a consequence of the hydrophobic effect. This means that proteins tend to bury hydroph ...
Transcription
... one of 3 stop codons ( UAG , UAA , UGA ) appears in A-site of the ribosome . A protein called release factor recognize stop codons and hydrolysis the bond between the last tRNA at the P-site and the polypeptide releasing them . The ribosomal subunits ...
... one of 3 stop codons ( UAG , UAA , UGA ) appears in A-site of the ribosome . A protein called release factor recognize stop codons and hydrolysis the bond between the last tRNA at the P-site and the polypeptide releasing them . The ribosomal subunits ...
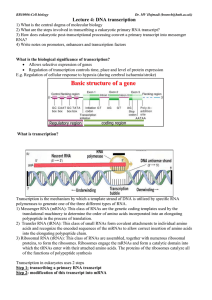
Lecture 4: DNA transcription
... Poly(A)polymerase and cleavage & polyadenylation specificity factor (CPSF) attach poly(A) generated from ATP ...
... Poly(A)polymerase and cleavage & polyadenylation specificity factor (CPSF) attach poly(A) generated from ATP ...
Lecture 11
... 3. The allosteric regulators bind to sites that are not active sites and elicit their effects by causing a Change in Shape of the Catalytic Subunit ...
... 3. The allosteric regulators bind to sites that are not active sites and elicit their effects by causing a Change in Shape of the Catalytic Subunit ...
Document
... DNA level, which corresponded to position of amino acid substitution in the gene product. Colinearity of mutations and altered amino acids in a subunit of tryptophan synthetase from E. coli C. Yanofsky, 1967. Scientific American ...
... DNA level, which corresponded to position of amino acid substitution in the gene product. Colinearity of mutations and altered amino acids in a subunit of tryptophan synthetase from E. coli C. Yanofsky, 1967. Scientific American ...
Name__________________________ Date______ Period
... 11. Where does translation occur in a cell? 12. The cell organelle known as the ___________ is where proteins are made. 13. Amino acids are carried to the ribosome by ___________. 14. Transfer RNA (tRNA) has a sequence of three nucleotides called the _____________ that binds to the ________ of mRNA. ...
... 11. Where does translation occur in a cell? 12. The cell organelle known as the ___________ is where proteins are made. 13. Amino acids are carried to the ribosome by ___________. 14. Transfer RNA (tRNA) has a sequence of three nucleotides called the _____________ that binds to the ________ of mRNA. ...
medmicro4-weapons delivery – G+
... 4. ‘Sorting’ via C-terminal wall-associating signals Vast majority of Gram + wall-associated proteins share structurally similar C-terminal wall-associating signals ...
... 4. ‘Sorting’ via C-terminal wall-associating signals Vast majority of Gram + wall-associated proteins share structurally similar C-terminal wall-associating signals ...
Electrontransfer proteins
... Electrontransfer component of the sulphur-bacteria. 2. Plant-type ferredoxin: [FeIII2(S2−)2]2+(RS−)4 In resting state 2 FeIII, but only one of them is reduced to FeII FeII−FeIII (1 unpaired electron, significant Fe-Fe interaction, but not complete electron delocalisation ...
... Electrontransfer component of the sulphur-bacteria. 2. Plant-type ferredoxin: [FeIII2(S2−)2]2+(RS−)4 In resting state 2 FeIII, but only one of them is reduced to FeII FeII−FeIII (1 unpaired electron, significant Fe-Fe interaction, but not complete electron delocalisation ...
Protein Metabolism
... stabilizing residue such as methionine or proline does not. – E3 enzymes are the readers of N-terminal residues. ...
... stabilizing residue such as methionine or proline does not. – E3 enzymes are the readers of N-terminal residues. ...
martakmalina proteins
... proportions of regular secondary structures such as α-helix or β-pleated sheet As a consequence of their rodlike or sheetlike shapes, these proteins exhibit fibrous character and have structural rather than dynamic roles Examples of fibrous proteins include keratin ...
... proportions of regular secondary structures such as α-helix or β-pleated sheet As a consequence of their rodlike or sheetlike shapes, these proteins exhibit fibrous character and have structural rather than dynamic roles Examples of fibrous proteins include keratin ...
Protein Synthesis
... 1. The amino acids coded for by the codons are linked together to make a protein. 2. Location: ribosome 3. mRNA travels out of the nucleus to a ribosome. ...
... 1. The amino acids coded for by the codons are linked together to make a protein. 2. Location: ribosome 3. mRNA travels out of the nucleus to a ribosome. ...
SR protein

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are ""S"" and ""R"" respectively. SR proteins are 50-300 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS binding domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.SR proteins were discovered in the 1990s in Drosophila and in amphibian oocytes, and later in humans. In general, metazoans appear to have SR proteins and unicellular organisms lack SR proteins.SR proteins are important in constitutive and alternative pre-mRNA splicing, mRNA export, genome stabilization, nonsense-mediated decay, and translation. SR proteins alternatively splice pre-mRNA by preferentially selecting different splice sites on the pre-mRNA strands to create multiple mRNA transcripts from one pre-mRNA transcript. Once splicing is complete the SR protein may or may not remain attached to help shuttle the mRNA strand out of the nucleus. As RNA Polymerase II is transcribing DNA into RNA, SR proteins attach to newly made pre-mRNA to prevent the pre-mRNA from binding to the coding DNA strand to increase genome stabilization. Topoisomerase I and SR proteins also interact to increase genome stabilization. SR proteins can control the concentrations of specific mRNA that is successfully translated into protein by selecting for nonsense-mediated decay codons during alternative splicing. SR proteins can alternatively splice NMD codons into its own mRNA transcript to auto-regulate the concentration of SR proteins. Through the mTOR pathway and interactions with polyribosomes, SR proteins can increase translation of mRNA.Ataxia telangiectasia, neurofibromatosis type 1, several cancers, HIV-1, and spinal muscular atrophy have all been linked to alternative splicing by SR proteins.