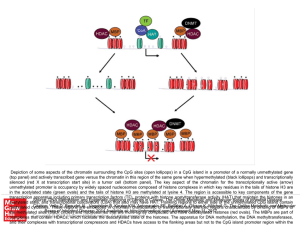
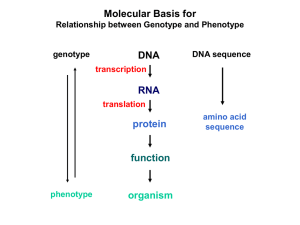
CS 262—Lecture 1 Notes • 4-‐5 HWs, 3 late days • (Optional
... • DNA must be read from 5’ to 3’ end • RNA usually single-‐stranded o Uracil replaces Thymine in RNA • Gene transcription: Transcription factors recognize binding sites in DNA, recruits RNA polymerase o RNA ...
... • DNA must be read from 5’ to 3’ end • RNA usually single-‐stranded o Uracil replaces Thymine in RNA • Gene transcription: Transcription factors recognize binding sites in DNA, recruits RNA polymerase o RNA ...
Eukaryotic Gene Control
... bodies): • Highly methylated (-CH3) bases, particularly cytosine • Removing of methyl groups can activate these genes ...
... bodies): • Highly methylated (-CH3) bases, particularly cytosine • Removing of methyl groups can activate these genes ...
Transcription factors - Raleigh Charter High School
... • Enhancer - specific DNA sequences which bind with activators to enhance transcription. • Activator - transcription factor which binds to an enhancer and stimulates transcription of gene. help position of the initiation complex on the promoter. • TATA Box - the DNA sequence which indicates where th ...
... • Enhancer - specific DNA sequences which bind with activators to enhance transcription. • Activator - transcription factor which binds to an enhancer and stimulates transcription of gene. help position of the initiation complex on the promoter. • TATA Box - the DNA sequence which indicates where th ...
Slide 1 - Ommbid.com
... (top panel) and actively transcribed gene versus the chromatin in this region of the same gene when hypermethylated (black lollipops) and transcriptionally silenced (red X at transcription start site) in a tumor cell (bottom panel). The key aspect of the chromatin for the transcriptionally active (a ...
... (top panel) and actively transcribed gene versus the chromatin in this region of the same gene when hypermethylated (black lollipops) and transcriptionally silenced (red X at transcription start site) in a tumor cell (bottom panel). The key aspect of the chromatin for the transcriptionally active (a ...
Distinguish between these 3 root types: - mvhs
... beginning of each gene that bind to ___________________ to begin transcription. ...
... beginning of each gene that bind to ___________________ to begin transcription. ...
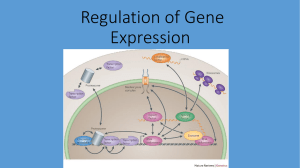
Regulation of Gene Expression
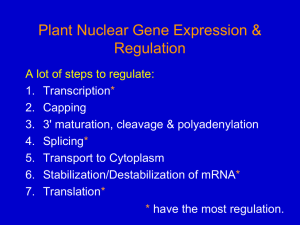
... transcription of mRNA from the gene that codes for the polypeptide • Post-transcriptional Control: the cell may transcribe the mRNA but break it down before translation. It can also modify the poly-A tail. WHY? • Post-translational Control: after synthesizing the polypeptide the cell can modify it c ...
... transcription of mRNA from the gene that codes for the polypeptide • Post-transcriptional Control: the cell may transcribe the mRNA but break it down before translation. It can also modify the poly-A tail. WHY? • Post-translational Control: after synthesizing the polypeptide the cell can modify it c ...
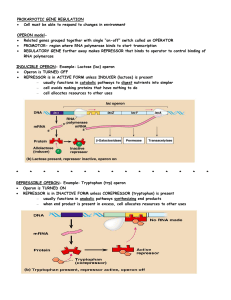
PROKARYOTIC GENE REGULATION
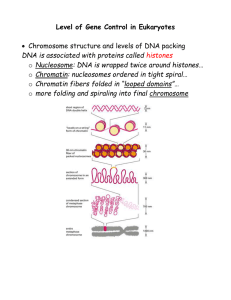
... LEVELS OF DNA PACKING REGULATE GENE EXPRESSION DNA packaged in chromatin fibers- regulates access to DNA by RNA ...
... LEVELS OF DNA PACKING REGULATE GENE EXPRESSION DNA packaged in chromatin fibers- regulates access to DNA by RNA ...
DNA Function II - Complete Vocab with
... General Transcription Factors: Other enzymes/proteins that are required for RNA Polymerase to function Transcription Activators: Proteins that bind to enhancers to stimulate transcription Transcription Repressors: Proteins that bind to enhancers to shut down transcription Enhancer: A sequence of DNA ...
... General Transcription Factors: Other enzymes/proteins that are required for RNA Polymerase to function Transcription Activators: Proteins that bind to enhancers to stimulate transcription Transcription Repressors: Proteins that bind to enhancers to shut down transcription Enhancer: A sequence of DNA ...
Regulation of Gene Expression
... – Remember that a promoter is where RNA polymerase binds to DNA to begin transcription – Occurs in prokaryotic genomes ...
... – Remember that a promoter is where RNA polymerase binds to DNA to begin transcription – Occurs in prokaryotic genomes ...
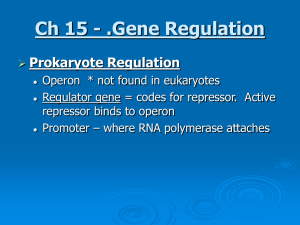
Ch 15 - .Gene Regulation
... 2** – Transcriptional control – [transcription factors- proteins that initiate RNA pol. binding] once a gene is unpackaged it will be transcribed. (transposons shut down genes by interrupting code, gene jumping, also encourages mutations) ...
... 2** – Transcriptional control – [transcription factors- proteins that initiate RNA pol. binding] once a gene is unpackaged it will be transcribed. (transposons shut down genes by interrupting code, gene jumping, also encourages mutations) ...
04/03
... Both enhancers and silencers affect transcription rate. Each has unique DNA sequence for the binding of regulatory proteins. Enhancer sequences contain multiple binding sites for trans-acting regulatory proteins. Enhancers could be located upstream from the promoter, downstream from the gene, or eve ...
... Both enhancers and silencers affect transcription rate. Each has unique DNA sequence for the binding of regulatory proteins. Enhancer sequences contain multiple binding sites for trans-acting regulatory proteins. Enhancers could be located upstream from the promoter, downstream from the gene, or eve ...
Table S11 Properties of the transcription factors for which binding
... bZIP transcription factor (ATF/CREB1 homolog) that regulates the unfolded protein response, via UPRE binding, and membrane biogenesis; ER stress-induced splicing pathway utilizing Ire1p, Trl1p and Ada5p facilitates efficient Hac1p synthesis ...
... bZIP transcription factor (ATF/CREB1 homolog) that regulates the unfolded protein response, via UPRE binding, and membrane biogenesis; ER stress-induced splicing pathway utilizing Ire1p, Trl1p and Ada5p facilitates efficient Hac1p synthesis ...
Lecture 7
... average are needed per amino acid in a mature protein), there is a real premium placed on the efficient production of proteins. Thus, the cell carefully decides which protein to produce and how much of that protein to produce. At the level of RNA production, this means that the genes being transcrib ...
... average are needed per amino acid in a mature protein), there is a real premium placed on the efficient production of proteins. Thus, the cell carefully decides which protein to produce and how much of that protein to produce. At the level of RNA production, this means that the genes being transcrib ...
Nuclear gene expression 1
... 1. Enhancers stimulate transcription, while Silencers inhibit. 2. Orientation-independent – Flip 180 degrees, still work 3. Position-independent (mostly) – Can work at a distance from promoter core – Enhancers have been found all over 4. Bind regulatory transcription factors ...
... 1. Enhancers stimulate transcription, while Silencers inhibit. 2. Orientation-independent – Flip 180 degrees, still work 3. Position-independent (mostly) – Can work at a distance from promoter core – Enhancers have been found all over 4. Bind regulatory transcription factors ...
A CAAT–Box Binding Factor Gene That Regulates Seed Development
... •Transcription factors are sequence-specific DNA binding factors proteins. They promote or block transcription by controlling the recruitment of RNA polymerase •Transcription is initiated at regions of DNA called promoters. Specific sequences of nucleotide bases at a promoter are recognized by both ...
... •Transcription factors are sequence-specific DNA binding factors proteins. They promote or block transcription by controlling the recruitment of RNA polymerase •Transcription is initiated at regions of DNA called promoters. Specific sequences of nucleotide bases at a promoter are recognized by both ...
24 October - web.biosci.utexas.edu
... discussion sections or on next Monday's class no later than 12:00PM. Email attachments and late delivery are not acceptable. 1. What factors ensure the fidelity of replication during DNA synthesis? 2. Define “promoter” and discuss the common features of bacterial promoters. 3. Describe functions of ...
... discussion sections or on next Monday's class no later than 12:00PM. Email attachments and late delivery are not acceptable. 1. What factors ensure the fidelity of replication during DNA synthesis? 2. Define “promoter” and discuss the common features of bacterial promoters. 3. Describe functions of ...
Study Guide MBMB 451A Fall 2002
... and IIH. What are TAFs and are they important for basal transcription. 7. What is an enhancer? What is a response element? 8. Describe two models for how an enhancer could effect the level of transcription. 9. What are the transcription factors called that are used by Pol I and Pol III? 10. Discuss ...
... and IIH. What are TAFs and are they important for basal transcription. 7. What is an enhancer? What is a response element? 8. Describe two models for how an enhancer could effect the level of transcription. 9. What are the transcription factors called that are used by Pol I and Pol III? 10. Discuss ...
LOYOLA COLLEGE (AUTONOMOUS), CHENNAI – 600 034
... 4. Define DNA methylation. 5. List out the functions of UTRs. 6. What are Tumour suppressor genes? Give examples. 7. What are non -viral retro-transposons? 8. What is the importance of HMG proteins in nucleosome organization? 9. Define Transcription attenuation. 10. Comment on RNA life span. PART B ...
... 4. Define DNA methylation. 5. List out the functions of UTRs. 6. What are Tumour suppressor genes? Give examples. 7. What are non -viral retro-transposons? 8. What is the importance of HMG proteins in nucleosome organization? 9. Define Transcription attenuation. 10. Comment on RNA life span. PART B ...
Genetic Controls in Eukaryotes
... Regulation at post-transcriptional level - RNA processing o Alternative RNA splicing = different segments of RNA are treated as exons and introns = different mRNA o Controlled by regulatory proteins specific to each cell type o Consequence = a single gene can code for more than one polypeptide = ...
... Regulation at post-transcriptional level - RNA processing o Alternative RNA splicing = different segments of RNA are treated as exons and introns = different mRNA o Controlled by regulatory proteins specific to each cell type o Consequence = a single gene can code for more than one polypeptide = ...
MSc / BSc positions in Systems Biology of Gene Regulation
... What defines the identity of a cell? How is the same genetic code used to build more than 200 different cell types with distinct physiological and morphological properties? These fundamental questions drive our enthusiasm for understanding how information processing is regulated at the level of chro ...
... What defines the identity of a cell? How is the same genetic code used to build more than 200 different cell types with distinct physiological and morphological properties? These fundamental questions drive our enthusiasm for understanding how information processing is regulated at the level of chro ...
Control of gene expression in prokaryotes and eukaryotes
... Gene expression is transcription of DNA to make RNA and then using the RNA to make proteins. This process can’t be left on indefinitely. The turning on and off of genes is critical to the development of an organism and the organism functioning properly throughout its life. Eukaryotic control Pretran ...
... Gene expression is transcription of DNA to make RNA and then using the RNA to make proteins. This process can’t be left on indefinitely. The turning on and off of genes is critical to the development of an organism and the organism functioning properly throughout its life. Eukaryotic control Pretran ...
«題目»
... Abstract The molecular basis of epigenetics involves modifications to DNA and histone proteins that associate with the regulation of gene expression but that do not result from mutation or changes to the DNA sequence. The four core histone proteins are subject to post-translational modifications, su ...
... Abstract The molecular basis of epigenetics involves modifications to DNA and histone proteins that associate with the regulation of gene expression but that do not result from mutation or changes to the DNA sequence. The four core histone proteins are subject to post-translational modifications, su ...
PartFourSumm_ThemesInRegulation.doc
... some enhancers can work by increasing the probability that a gene is in a permissive environment for transcription (putative accessible chromatin). b. Much evidence has implicated enzymes such as histone acetyl transferases (HATs, e.g. protein complexes containing Gcn5p + Ada2 in yeast, PCAF + P300/ ...
... some enhancers can work by increasing the probability that a gene is in a permissive environment for transcription (putative accessible chromatin). b. Much evidence has implicated enzymes such as histone acetyl transferases (HATs, e.g. protein complexes containing Gcn5p + Ada2 in yeast, PCAF + P300/ ...