Document
... • Precise subsequence can be selected using appropriate primers • Can create large amounts from small sample • Sine qua none for DNA sequencing projects, and a lot of experimental biology Lecture 1 CS566 ...
... • Precise subsequence can be selected using appropriate primers • Can create large amounts from small sample • Sine qua none for DNA sequencing projects, and a lot of experimental biology Lecture 1 CS566 ...
Module 4 PowerPoint Slides - The Cancer 101 Curriculum
... Most disease begin in our genes. If DNA repair fails in a cell, mutations can be passed on to future copies. Gene mutations can have a latent effect, or even a positive effect. The ability to identify a gene mutation is possible through genetic testing. ...
... Most disease begin in our genes. If DNA repair fails in a cell, mutations can be passed on to future copies. Gene mutations can have a latent effect, or even a positive effect. The ability to identify a gene mutation is possible through genetic testing. ...
Human Genome Structure and Organization
... • Encode proteins (and some RNAs) • Human genetics is the study of gene variation in humans • ‘Gene’ as a term is used ambiguously to refer both to the ‘locus’ and the ‘allele’ ie- There is only one locus but two alleles in a given individual. • Sequencing in both genome projects took place upon mul ...
... • Encode proteins (and some RNAs) • Human genetics is the study of gene variation in humans • ‘Gene’ as a term is used ambiguously to refer both to the ‘locus’ and the ‘allele’ ie- There is only one locus but two alleles in a given individual. • Sequencing in both genome projects took place upon mul ...
Population Genetics Sequence Diversity Molecular Evolution
... Homo sapiens A powerful idea: We are a very young species. 1 - 1.5 million years old but population bottleneck 200,000 years ago. We are closely related 10,000 meiosis = 200,000 years In Great Britain estimates predict that two unrelated people share common ancestor not more than 22 generations ago ...
... Homo sapiens A powerful idea: We are a very young species. 1 - 1.5 million years old but population bottleneck 200,000 years ago. We are closely related 10,000 meiosis = 200,000 years In Great Britain estimates predict that two unrelated people share common ancestor not more than 22 generations ago ...
No Slide Title
... anode (indium tin oxide-coated polyethylene terephthalate) and power generation under light and dark conditions was evaluated using a single-chamber bio-photovoltaic cell (BPV) system. Increased power outputs were observed for all strains upon illumination, with the largest light effect observed for ...
... anode (indium tin oxide-coated polyethylene terephthalate) and power generation under light and dark conditions was evaluated using a single-chamber bio-photovoltaic cell (BPV) system. Increased power outputs were observed for all strains upon illumination, with the largest light effect observed for ...
PERSONALIZED MEDICINE Health Care Focus for the Future
... If shared mutation association identified, then search undiagnosed patients for the identical mutation to confirm disease diagnosis ...
... If shared mutation association identified, then search undiagnosed patients for the identical mutation to confirm disease diagnosis ...
TOC - G3: Genes | Genomes | Genetics
... Additionally, the authors found that duplicate cycling genes preferentially retain the same or similar timing of cyclic expression and that this timing may be controlled, in part, by putative cis-regulatory elements. ...
... Additionally, the authors found that duplicate cycling genes preferentially retain the same or similar timing of cyclic expression and that this timing may be controlled, in part, by putative cis-regulatory elements. ...
ppt - Sol Genomics Network
... - TIGR full-length cDNA sequences (redundantly sequenced) - SGN unigene contigs with 5 or more ESTs - redundnacy correction 456 of 8,097 genes found in available genome sequence (5.6%) Correcting for 85% expectation yields 6.6% of target gene space 15.5/0.066 = 234 Mb tomato euchromatin target ...
... - TIGR full-length cDNA sequences (redundantly sequenced) - SGN unigene contigs with 5 or more ESTs - redundnacy correction 456 of 8,097 genes found in available genome sequence (5.6%) Correcting for 85% expectation yields 6.6% of target gene space 15.5/0.066 = 234 Mb tomato euchromatin target ...
Human Genome Project
... • The human genome's gene-dense "urban centers" are predominantly composed of the DNA building blocks G and C. • In contrast, the gene-poor "deserts" are rich in the DNA building blocks A and T. GC- and AT-rich regions usually can be seen through a microscope as light and dark bands on chromosomes. ...
... • The human genome's gene-dense "urban centers" are predominantly composed of the DNA building blocks G and C. • In contrast, the gene-poor "deserts" are rich in the DNA building blocks A and T. GC- and AT-rich regions usually can be seen through a microscope as light and dark bands on chromosomes. ...
Document
... • The human genome's gene-dense "urban centers" are predominantly composed of the DNA building blocks G and C. • In contrast, the gene-poor "deserts" are rich in the DNA building blocks A and T. GC- and AT-rich regions usually can be seen through a microscope as light and dark bands on chromosomes. ...
... • The human genome's gene-dense "urban centers" are predominantly composed of the DNA building blocks G and C. • In contrast, the gene-poor "deserts" are rich in the DNA building blocks A and T. GC- and AT-rich regions usually can be seen through a microscope as light and dark bands on chromosomes. ...
a version - SEA
... In the fall semester we isolated six bacteriophages from Gordonia terrae and two phages from Mycobacterium smegmatis. We found, through transmission electron microscopy, that the phages all had siphoviral morphology. Because there have been a plethora of mycobacteriophages sequenced, we submitted tw ...
... In the fall semester we isolated six bacteriophages from Gordonia terrae and two phages from Mycobacterium smegmatis. We found, through transmission electron microscopy, that the phages all had siphoviral morphology. Because there have been a plethora of mycobacteriophages sequenced, we submitted tw ...
Comparative Genome Organization in plants: From Sequence and Markers to... and Chromosomes Summary
... connected by linker DNA. Repetitive sequences probably play a key role in stabilizing this structure. Chromatin Remodeling and Histone Acetylation: Histone acetylation is known to change the structure of the chromatin. It does it by modulating the position of nucleosomes. Changes in nucleosome posit ...
... connected by linker DNA. Repetitive sequences probably play a key role in stabilizing this structure. Chromatin Remodeling and Histone Acetylation: Histone acetylation is known to change the structure of the chromatin. It does it by modulating the position of nucleosomes. Changes in nucleosome posit ...
SEGMENTAL VARIATION
... • Depth of coverage in a single patient was compared to average and standard deviation of depth of coverage. • Algorithms were developed for: – Classifying X chromosome as being deleted in males compared with females – Classifying X chromosome as being duplicated in females compared with males S L I ...
... • Depth of coverage in a single patient was compared to average and standard deviation of depth of coverage. • Algorithms were developed for: – Classifying X chromosome as being deleted in males compared with females – Classifying X chromosome as being duplicated in females compared with males S L I ...
Genomics
... time preserving the environment. Genomics is an entry point for looking at the other ‘omics’ sciences. The information in the genes of an organism, its genotype, is largely responsible for the final physical makeup of the organism, referred to as the “phenotype”. However, the environment also has so ...
... time preserving the environment. Genomics is an entry point for looking at the other ‘omics’ sciences. The information in the genes of an organism, its genotype, is largely responsible for the final physical makeup of the organism, referred to as the “phenotype”. However, the environment also has so ...
Lecture_4
... be able to predict function - NOT assign function. – The biological function of many genes have not been determined, even in model systems. – As genomic characterization of gene function continues - more and more computer generated annotations will be correct. ...
... be able to predict function - NOT assign function. – The biological function of many genes have not been determined, even in model systems. – As genomic characterization of gene function continues - more and more computer generated annotations will be correct. ...
SNP Discovery by sequencing 1000 genomes
... searched for the genetic cause for > 25 years…….. Late last year, he finally found it-by sequencing his entire genome -in SH3TC2 (the SH3 domain and tetratricopeptide repeats 2 gene) – cost ~$50,000 First to show how whole-genome sequencing can be used to identify the genetic cause of an individual' ...
... searched for the genetic cause for > 25 years…….. Late last year, he finally found it-by sequencing his entire genome -in SH3TC2 (the SH3 domain and tetratricopeptide repeats 2 gene) – cost ~$50,000 First to show how whole-genome sequencing can be used to identify the genetic cause of an individual' ...
Using Data from the Human Genome Project in
... Don't let that scare you away. As usage of the sites has increased, the labs that maintain these pages have made them easier to use and understand. They've now reached a point where they can be used in an intuitive way by most people with a basic understanding of molecular biology. I've written a br ...
... Don't let that scare you away. As usage of the sites has increased, the labs that maintain these pages have made them easier to use and understand. They've now reached a point where they can be used in an intuitive way by most people with a basic understanding of molecular biology. I've written a br ...
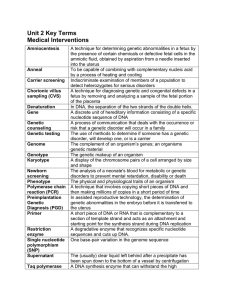
Unit 2 Terms
... In DNA, the separation of the two strands of the double helix. A discrete unit of hereditary information consisting of a specific nucleotide sequence of DNA A process of communication that deals with the occurrence or risk that a genetic disorder will occur in a family The use of methods to determin ...
... In DNA, the separation of the two strands of the double helix. A discrete unit of hereditary information consisting of a specific nucleotide sequence of DNA A process of communication that deals with the occurrence or risk that a genetic disorder will occur in a family The use of methods to determin ...
Group 4 members
... – High throughput deep sequencing analyze pools of cells, get genome-wide overviews of genes and enable rapid assessment of the spectrum of genes, assigning genes to phenotypes with high saturation and accuracy; ...
... – High throughput deep sequencing analyze pools of cells, get genome-wide overviews of genes and enable rapid assessment of the spectrum of genes, assigning genes to phenotypes with high saturation and accuracy; ...
A Next Generation Sequencing Panel for DNA Typing of
... as mixtures are commonly seen in forensic analysis, it would be beneficial if determination as well as quantification of mixture components can be performed. Therefore, we have designed and evaluated a new NGS panel for analyses of severely degraded or mixed DNA samples. The final goal is to optimis ...
... as mixtures are commonly seen in forensic analysis, it would be beneficial if determination as well as quantification of mixture components can be performed. Therefore, we have designed and evaluated a new NGS panel for analyses of severely degraded or mixed DNA samples. The final goal is to optimis ...
9 Genomics and Beyond
... each piece is cloned, forming a DNA library. (2) The DNA fragments must overlap other fragments, so the restriction enzyme is not allowed to cut at every possible restriction site. (3) Computers assemble the fragments into contigs by determining which DNA pieces have bands that are common. (4) Uniqu ...
... each piece is cloned, forming a DNA library. (2) The DNA fragments must overlap other fragments, so the restriction enzyme is not allowed to cut at every possible restriction site. (3) Computers assemble the fragments into contigs by determining which DNA pieces have bands that are common. (4) Uniqu ...
Clinical genomics - University of Toledo
... • The number, indications, and complexity of genetic tests offered have been increasing, and will continue to do so for the foreseeable future. • It is therefore not surprising that mistakes often occur in the ordering of complex genetic tests. • Incorrect ordering of genetic tests results in unnece ...
... • The number, indications, and complexity of genetic tests offered have been increasing, and will continue to do so for the foreseeable future. • It is therefore not surprising that mistakes often occur in the ordering of complex genetic tests. • Incorrect ordering of genetic tests results in unnece ...
Whole genome sequencing

Whole genome sequencing (also known as full genome sequencing, complete genome sequencing, or entire genome sequencing) is a laboratory process that determines the complete DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast.Whole genome sequencing should not be confused with DNA profiling, which only determines the likelihood that genetic material came from a particular individual or group, and does not contain additional information on genetic relationships, origin or susceptibility to specific diseases. Also unlike full genome sequencing, SNP genotyping covers less than 0.1% of the genome. Almost all truly complete genomes are of microbes; the term ""full genome"" is thus sometimes used loosely to mean ""greater than 95%"". The remainder of this article focuses on nearly complete human genomes.High-throughput genome sequencing technologies have largely been used as a research tool and are currently being introduced in the clinics. In the future of personalized medicine, whole genome sequence data will be an important tool to guide therapeutic intervention. The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response.