Microbial Genomics
... is hybridized to the target DNA. RED represents Sample DNA where either DNA or cDNA is derived from diseased tissue hybridized to the target DNA. YELLOW represents a combination of Control and Sample DNA where both hybridized equally to the target DNA. BLACK represents areas where neither the Contro ...
... is hybridized to the target DNA. RED represents Sample DNA where either DNA or cDNA is derived from diseased tissue hybridized to the target DNA. YELLOW represents a combination of Control and Sample DNA where both hybridized equally to the target DNA. BLACK represents areas where neither the Contro ...
FILTUS
... It is revealed to us that the parents of our patient are first cousins. This suggests that the causal variant lies in an autozygous stretch of the genome – i.e. a long homozygous region where both haplotypes originate from the same great grandparent. Restricting our search to these regions will hope ...
... It is revealed to us that the parents of our patient are first cousins. This suggests that the causal variant lies in an autozygous stretch of the genome – i.e. a long homozygous region where both haplotypes originate from the same great grandparent. Restricting our search to these regions will hope ...
Supplemental Figure 3
... representative A. thaliana acccessions. Equivalent amounts of genomic DNA isolated from different accessions were subjected to PCR using the same pair of SCR1 primers (the PseSCR3 and PseSCR5 primers described by Shimizu et al. 2004). Note that DNA from the C24 and Mt-0 accessions, which lack SCR1 ...
... representative A. thaliana acccessions. Equivalent amounts of genomic DNA isolated from different accessions were subjected to PCR using the same pair of SCR1 primers (the PseSCR3 and PseSCR5 primers described by Shimizu et al. 2004). Note that DNA from the C24 and Mt-0 accessions, which lack SCR1 ...
Slide 1
... Topics in CS262 Part 1: In-depth coverage of basic computational methods for analysis of biological sequences ...
... Topics in CS262 Part 1: In-depth coverage of basic computational methods for analysis of biological sequences ...
Cytogenetics to Cytogenomics: An Introduction to Genomic
... Arrays can be designed for wide coverage of the genome with specific applications. However, there are a few things that they cannot detect, such as balanced rearrangements.7 Although array technologies can detect variations at high resolution, new sequence variations cannot be detected, as prior kno ...
... Arrays can be designed for wide coverage of the genome with specific applications. However, there are a few things that they cannot detect, such as balanced rearrangements.7 Although array technologies can detect variations at high resolution, new sequence variations cannot be detected, as prior kno ...
DNA Sequencing and Gene Analysis
... • The four dNTPs are added one at a time, with apyrase degradation and washing in between. • The amount of light released is proportional to the number of bases added. Thus, if the sequence has 2 A’s in a row, both get added and twice as much light is released as would have happened with only 1 A. • ...
... • The four dNTPs are added one at a time, with apyrase degradation and washing in between. • The amount of light released is proportional to the number of bases added. Thus, if the sequence has 2 A’s in a row, both get added and twice as much light is released as would have happened with only 1 A. • ...
BASICS ON MOLECULAR BIOLOGY
... Base calling: identifying which base corresponds to each position in a read – Non-trivial problem! ...
... Base calling: identifying which base corresponds to each position in a read – Non-trivial problem! ...
The Human Genome Project - Homepages | The University of
... software was used to work out the "best fit" map of all the markers, with advanced statistical methods and algorithms ...
... software was used to work out the "best fit" map of all the markers, with advanced statistical methods and algorithms ...
View/print full test page
... genes in the panel plus ten bases into the introns and untranslated regions (5' and 3'). Sanger sequencing is performed to confirm variants suspected or confirmed to be pathogenic. o Deletion/duplication analysis is performed using a high resolution, custom microarray platform designed to target the ...
... genes in the panel plus ten bases into the introns and untranslated regions (5' and 3'). Sanger sequencing is performed to confirm variants suspected or confirmed to be pathogenic. o Deletion/duplication analysis is performed using a high resolution, custom microarray platform designed to target the ...
Case study: PacBio and Dovetail - For cashew genome, combining
... “There are lots of published plant genomes that are not very good quality because they were done exclusively with short-read sequencing, so there are lots of 'holes' in these Swiss-cheese-like genomes.” The scientists had previously worked with Single Molecule, Real-Time (SMRT®) Sequencing from PacB ...
... “There are lots of published plant genomes that are not very good quality because they were done exclusively with short-read sequencing, so there are lots of 'holes' in these Swiss-cheese-like genomes.” The scientists had previously worked with Single Molecule, Real-Time (SMRT®) Sequencing from PacB ...
1 Genomics 1. The world of “-omics” The field of “Genomics” has
... The Drosophila euchromatic genome is 120 Mb. This means 1.2 Gb were sequenced. With an average read length of ≈ 500 bp, there were ≈ 2.4 million sequences. Step 1: Pairwise comparison of each sequence to every other sequence to search for overlaps. The total number of pairwise comparisons is given b ...
... The Drosophila euchromatic genome is 120 Mb. This means 1.2 Gb were sequenced. With an average read length of ≈ 500 bp, there were ≈ 2.4 million sequences. Step 1: Pairwise comparison of each sequence to every other sequence to search for overlaps. The total number of pairwise comparisons is given b ...
Sequencing Medicago truncatula expressed sequenced tags
... learn that the authors want to know whether the protocol they describe can lead to the discovery of predicted genes not found in an existing database. For this question to make any sense, we have to know what the existing database is about. I presume that the 226,923 ESTs are the result of conventio ...
... learn that the authors want to know whether the protocol they describe can lead to the discovery of predicted genes not found in an existing database. For this question to make any sense, we have to know what the existing database is about. I presume that the 226,923 ESTs are the result of conventio ...
AP Biology 12 Chapter 21 Genomes and Their Evolution Enduring
... After the ancestors of humans and chimpanzees diverged as species, the fusion of two ancestral chromosomes in the human line led to different haploid numbers for humans (n = 23) and chimpanzees (n = 24). Another pattern with medical relevance was noted: The chromosomal breakage points associated wit ...
... After the ancestors of humans and chimpanzees diverged as species, the fusion of two ancestral chromosomes in the human line led to different haploid numbers for humans (n = 23) and chimpanzees (n = 24). Another pattern with medical relevance was noted: The chromosomal breakage points associated wit ...
Document
... - A single gene can code for multiple proteins using alternative splicing. Although all the DNA in a genome can be isolated from a single cell, only a portion of the proteome is expressed in a single cell or tissue. The transcriptome consists of all the RNA that is present in a cell or tissue. ...
... - A single gene can code for multiple proteins using alternative splicing. Although all the DNA in a genome can be isolated from a single cell, only a portion of the proteome is expressed in a single cell or tissue. The transcriptome consists of all the RNA that is present in a cell or tissue. ...
Chalmers_Bioinformatics
... • DNA is generally immobilized on a solid support • Very large numbers of small reads • Multiple reads of a each section of genomic DNA (eg 30x) • Assembling the genome becomes a significant computational problem • Some ‘single molecule’ methods do not require PCR (reduces errors) • Cost has reduced ...
... • DNA is generally immobilized on a solid support • Very large numbers of small reads • Multiple reads of a each section of genomic DNA (eg 30x) • Assembling the genome becomes a significant computational problem • Some ‘single molecule’ methods do not require PCR (reduces errors) • Cost has reduced ...
Freshman Seminar
... The biological importance of sequence alignment • Sequence alignments assess the degree of similarity between sequences • Similar sequences suggest similar function – Proteins with similar sequences are likely to play similar biochemical roles – Regulatory DNA sequences that are similar will likely ...
... The biological importance of sequence alignment • Sequence alignments assess the degree of similarity between sequences • Similar sequences suggest similar function – Proteins with similar sequences are likely to play similar biochemical roles – Regulatory DNA sequences that are similar will likely ...
additional file s4 - Springer Static Content Server
... of Polyvinylpyrrolidone (PVP 40 000) was added to buffer AP1. Among the nine individuals previously studied in Rønsted et al. {Rønsted, 2007 #45}, we extracted DNA of two according to this protocol. For the remaining seven samples, we used total genomic DNA of already extracted these authors. Amplif ...
... of Polyvinylpyrrolidone (PVP 40 000) was added to buffer AP1. Among the nine individuals previously studied in Rønsted et al. {Rønsted, 2007 #45}, we extracted DNA of two according to this protocol. For the remaining seven samples, we used total genomic DNA of already extracted these authors. Amplif ...
A general video on DNA sequencing is
... a) Explain why a gene chip (i.e. a DNA Microarray) would be ideal to use when determining which genes are being turned on (i.e. proteins expressed) and which genes are being turned off during cell division (or any other cell process). b) You have isolated the mRNA from cancerous tissue and labeled i ...
... a) Explain why a gene chip (i.e. a DNA Microarray) would be ideal to use when determining which genes are being turned on (i.e. proteins expressed) and which genes are being turned off during cell division (or any other cell process). b) You have isolated the mRNA from cancerous tissue and labeled i ...
Interpretive Criteria for Identification of Bacteria and
... identification of organisms. Highly conserved genes, such as the ribosomal RNA (rRNA) genes, provide information on the general properties of organisms based on the properties of their known relatives. Other gene sequences can provide further detail, eg, as may be needed to distinguish a pathogen fr ...
... identification of organisms. Highly conserved genes, such as the ribosomal RNA (rRNA) genes, provide information on the general properties of organisms based on the properties of their known relatives. Other gene sequences can provide further detail, eg, as may be needed to distinguish a pathogen fr ...
Journal Club - Clinical Chemistry
... Figure 1. Detection of microsatellite instability by MSI-PCR and next-generation DNA sequencing. Representative capillary electrophoresis results from MSI-PCR (left) and “virtual electropherograms” of next-generation DNA sequencing data (right). The length (x-axis) and relative abundance (Y-axis) of ...
... Figure 1. Detection of microsatellite instability by MSI-PCR and next-generation DNA sequencing. Representative capillary electrophoresis results from MSI-PCR (left) and “virtual electropherograms” of next-generation DNA sequencing data (right). The length (x-axis) and relative abundance (Y-axis) of ...
Are My Genes Mutated? Analyzing Loss of Function Variants in the
... • These variants were found to: - contain stop codons, disrupt splice-sites, or result in insertions or deletions that change DNA reading frame - primarily be enriched in low-frequency alleles; those present in higher frequencies are either present in poorly evolutionary conserved genes, multigene f ...
... • These variants were found to: - contain stop codons, disrupt splice-sites, or result in insertions or deletions that change DNA reading frame - primarily be enriched in low-frequency alleles; those present in higher frequencies are either present in poorly evolutionary conserved genes, multigene f ...
Genomics and museum specimens - Integrative Biology
... Genome-wide surveys of genetic variation in museum and present-day samples should thus enable us to identify loci that have responded to selection, including particular environmental changes. The work conducted by Wilson and colleagues nearly 25 years ago and the study by Bi et al. (2013) were both ...
... Genome-wide surveys of genetic variation in museum and present-day samples should thus enable us to identify loci that have responded to selection, including particular environmental changes. The work conducted by Wilson and colleagues nearly 25 years ago and the study by Bi et al. (2013) were both ...
The Human Genome Project
... region of the genome, produce a virtually unlimited number of copies of it, and determine its nucleotide sequence overnight. • At the height of the Human Genome Project, sequencing factories were generating DNA sequences at a rate of 1000 nucleotides per second 24/7. • Technical breakthroughs that a ...
... region of the genome, produce a virtually unlimited number of copies of it, and determine its nucleotide sequence overnight. • At the height of the Human Genome Project, sequencing factories were generating DNA sequences at a rate of 1000 nucleotides per second 24/7. • Technical breakthroughs that a ...
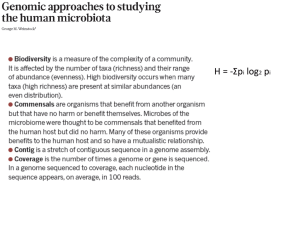
Slide 1
... Antibiotic treatment perturbs the microbiome by reducing its size and altering its composition. This disturbance can lead to infection, and antibiotic-resistant organisms such as Clostridium difficile — normally controlled by the microbiome — can overgrow and create problems. More complex community ...
... Antibiotic treatment perturbs the microbiome by reducing its size and altering its composition. This disturbance can lead to infection, and antibiotic-resistant organisms such as Clostridium difficile — normally controlled by the microbiome — can overgrow and create problems. More complex community ...
Call 2016, July: `GenOmics of rare diseases`
... development of diagnostic approaches and of innovative therapeutics. The recent development of massively parallel DNA sequencing technologies has provided a new and potentially powerful way to identify almost all the mutations responsible for Mendelian disorders. Whole exome sequenc ...
... development of diagnostic approaches and of innovative therapeutics. The recent development of massively parallel DNA sequencing technologies has provided a new and potentially powerful way to identify almost all the mutations responsible for Mendelian disorders. Whole exome sequenc ...
Exome sequencing

Exome sequencing is a technique for sequencing all the protein-coding genes in a genome (known as the exome). It consists of first selecting only the subset of DNA that encodes proteins (known as exons), and then sequencing that DNA using any high throughput DNA sequencing technology. There are 180,000 exons, which constitute about 1% of the human genome, or approximately 30 million base pairs, but mutations in these sequences are much more likely to have severe consequences than in the remaining 99%. The goal of this approach is to identify genetic variation that is responsible for both mendelian and common diseases such as Miller syndrome and Alzheimer's disease without the high costs associated with whole-genome sequencing.