The Possibilities and limitations of nucleic acid amphfication
... to 0.5- 10 ml for culture-is another important reason for false negative results. For DNA amplification reactions that start from a maximum sample volume of 50 p l (in a 100pl total reaction volume), it can be calculated that 20 DNA target moleculks (or organisms)/ml of sample is the detection limit ...
... to 0.5- 10 ml for culture-is another important reason for false negative results. For DNA amplification reactions that start from a maximum sample volume of 50 p l (in a 100pl total reaction volume), it can be calculated that 20 DNA target moleculks (or organisms)/ml of sample is the detection limit ...
Bacteria Screening PCR Kit
... pH8.0) then suspend in a 200 μl of Chelex® solution. 2) Heat at 99℃ for a period of 5 minutes then quickly cool by placing on ice for 1 minute or more. 3) Centrifuge at 12,000 rpm (approx. 13,000 X g ) for 1 minute. 4) Use the supernatant as the DNA Sample Solution for PCR. [ Option 2 ] (Use o ...
... pH8.0) then suspend in a 200 μl of Chelex® solution. 2) Heat at 99℃ for a period of 5 minutes then quickly cool by placing on ice for 1 minute or more. 3) Centrifuge at 12,000 rpm (approx. 13,000 X g ) for 1 minute. 4) Use the supernatant as the DNA Sample Solution for PCR. [ Option 2 ] (Use o ...
MARKER GENE TECHNOLOGIES, Inc
... We do not recommend reusing RedView precast gels as signal decreases with ...
... We do not recommend reusing RedView precast gels as signal decreases with ...
presentation - Genome-to-Genome Distance Calculator
... please find below the results of your GGDC job. A short explanation is found at the end of this email. Further information is provided on our website at http://www.gbdp.org/species/. If you use this service in a publication, please cite the appropriate references listed ...
... please find below the results of your GGDC job. A short explanation is found at the end of this email. Further information is provided on our website at http://www.gbdp.org/species/. If you use this service in a publication, please cite the appropriate references listed ...
Basic sequence analyses and submission
... Copy the M13_F sequence and click on RunVecScreen. Find the region corresponding to vector. Identify the Hind III cloning site: AAGCTT, and eliminate the vector sequence, but not the cloning site. Highlight the cloning site. Repeat the same process with the M13_R sequence. The other sequences (F1, F ...
... Copy the M13_F sequence and click on RunVecScreen. Find the region corresponding to vector. Identify the Hind III cloning site: AAGCTT, and eliminate the vector sequence, but not the cloning site. Highlight the cloning site. Repeat the same process with the M13_R sequence. The other sequences (F1, F ...
Amplicon metagenomics for elucidating microbial community
... The microbial composition of Solanum lycopersicum and Beta vulgaris has been previously studied by surveying their anatomical microbial ecology. Such work concluded that there are major differences in the microbial communities in different parts of the plant such as leaves, stem, fruits and roots an ...
... The microbial composition of Solanum lycopersicum and Beta vulgaris has been previously studied by surveying their anatomical microbial ecology. Such work concluded that there are major differences in the microbial communities in different parts of the plant such as leaves, stem, fruits and roots an ...
16_Lecture_Presentation
... Was found in the bacteria but not any of the radioLabeled protein because the bacteria “ingested” the viral DNA ...
... Was found in the bacteria but not any of the radioLabeled protein because the bacteria “ingested” the viral DNA ...
- Peanut Science
... labor. Finally, discovery of selfs typically occurs a year after the crosses are made, resulting in potential delays to improvement programs. Identification of hybrids can be performed through use of DNA markers. Codominant markers are preferable because they produce different alleles (markers) for ...
... labor. Finally, discovery of selfs typically occurs a year after the crosses are made, resulting in potential delays to improvement programs. Identification of hybrids can be performed through use of DNA markers. Codominant markers are preferable because they produce different alleles (markers) for ...
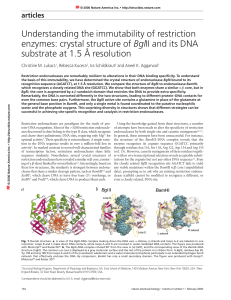
Article - School of Chemistry and Biochemistry
... of the central four strands of one monomer from each structure shows that the other monomers do not overlie, and are in fact displaced by as much as 4.3 Å in the outer helices. This alteration in the relationship of the monomers in the two structures leads to a narrower DNA binding cleft in BglII. B ...
... of the central four strands of one monomer from each structure shows that the other monomers do not overlie, and are in fact displaced by as much as 4.3 Å in the outer helices. This alteration in the relationship of the monomers in the two structures leads to a narrower DNA binding cleft in BglII. B ...
DNA is the genetic material
... DNA polymerases cannot initiate synthesis of a polynucleotide; they can only add nucleotides to the 3′ end The initial nucleotide chain is a short stretch of RNA called a primer, synthesized by the enzyme primase. Primase starts an RNA chain from a single RNA nucleotide, adding RNA nucleotides one a ...
... DNA polymerases cannot initiate synthesis of a polynucleotide; they can only add nucleotides to the 3′ end The initial nucleotide chain is a short stretch of RNA called a primer, synthesized by the enzyme primase. Primase starts an RNA chain from a single RNA nucleotide, adding RNA nucleotides one a ...
Use of a single primer to fluorescently label selective amplified
... combinations of selective primers (5). The use of fluorescently labeled selective primers in conjunction with a capillary sequencing device provides a robust platform for resolving fragment profiles, but a limiting factor for this type of resolution is the cost of fluorescently labeled selective pri ...
... combinations of selective primers (5). The use of fluorescently labeled selective primers in conjunction with a capillary sequencing device provides a robust platform for resolving fragment profiles, but a limiting factor for this type of resolution is the cost of fluorescently labeled selective pri ...
A physical map of the genome of Hmmophilus
... Digestion of DNA in agarose blocks. Usually digests were carried out on the DNA contained in one-third of a complete plug. Restriction einzyme buffers were diffused into the agarose blocks as outlined below. Plugs or portions of plugs were washed in Eppendorf tubes with 500 1.11 vlolumesof buffer (u ...
... Digestion of DNA in agarose blocks. Usually digests were carried out on the DNA contained in one-third of a complete plug. Restriction einzyme buffers were diffused into the agarose blocks as outlined below. Plugs or portions of plugs were washed in Eppendorf tubes with 500 1.11 vlolumesof buffer (u ...
Primary Sequence of Ovomucoid Messenger RNA as Determined
... was used to transform E. colf K strain RRI (4), as described in Materials and Methods. All bacterial transfers were carried out in a certified laminar flow hood in a P-3 physical containment facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines ...
... was used to transform E. colf K strain RRI (4), as described in Materials and Methods. All bacterial transfers were carried out in a certified laminar flow hood in a P-3 physical containment facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines ...
Primary Sequence of Ovomucoid Messenger RNA
... was used to transform E. colf K strain RRI (4), as described in Materials and Methods. All bacterial transfers were carried out in a certified laminar flow hood in a P-3 physical containment facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guideline ...
... was used to transform E. colf K strain RRI (4), as described in Materials and Methods. All bacterial transfers were carried out in a certified laminar flow hood in a P-3 physical containment facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guideline ...
A novel procedure for genotyping of single nucleotide polymorphisms in trisomy with genomic DNA and the invader assay.
... can accurately determine genotypes in trisomic DNA samples are expensive, require specialized equipment and complicated analyses. Here we report proof-of-concept results for an InvaderÕ assay-based genotyping procedure that can determine SNP genotypes in trisomic genomic DNA samples in a simple and ...
... can accurately determine genotypes in trisomic DNA samples are expensive, require specialized equipment and complicated analyses. Here we report proof-of-concept results for an InvaderÕ assay-based genotyping procedure that can determine SNP genotypes in trisomic genomic DNA samples in a simple and ...
Comparison between the efficiency of liposome and
... the reproducibility of the original SMGT [13]. Nevertheless, it becomes known to many researchers the obvious inefficiency of SMGT enhancement approaches to “integrate” the foreign DNA into the genome of the sperm [14]. An interesting molecular trick that represented by implicating restriction enzym ...
... the reproducibility of the original SMGT [13]. Nevertheless, it becomes known to many researchers the obvious inefficiency of SMGT enhancement approaches to “integrate” the foreign DNA into the genome of the sperm [14]. An interesting molecular trick that represented by implicating restriction enzym ...
Analysis of Two Genes Encoding Prothrombin Activators in
... The presence of two parallel prothrombin activators for an extremely different physiological role in the Pseudonaja textilis (P. textilis) snake is unique. One prothrombin activator, pseutarin C, is a multi subunit protein found in the venom as a toxin, and can activate blood clot in target preys. T ...
... The presence of two parallel prothrombin activators for an extremely different physiological role in the Pseudonaja textilis (P. textilis) snake is unique. One prothrombin activator, pseutarin C, is a multi subunit protein found in the venom as a toxin, and can activate blood clot in target preys. T ...
GBSA: a comprehensive software for analysing whole genome
... (MeDIP-seq) (3) or methyl-binding domain-isolated genome sequencing (MiGS) (4), MethylCap-seq (5) have been proven to be cost-effective approaches to study and compare differentially methylated regions in a variety of organisms, tissues and cells of several developmental stages. Meanwhile, the seque ...
... (MeDIP-seq) (3) or methyl-binding domain-isolated genome sequencing (MiGS) (4), MethylCap-seq (5) have been proven to be cost-effective approaches to study and compare differentially methylated regions in a variety of organisms, tissues and cells of several developmental stages. Meanwhile, the seque ...
Chapter 16 The Molecular Basis of Inheritance Multiple
... 5) After mixing a heat-killed, phosphorescent strain of bacteria with a living non-phosphorescent strain, you discover that some of the living cells are now phosphorescent. Which observations would provide the best evidence that the ability to fluoresce is a heritable trait? A) DNA passed from the ...
... 5) After mixing a heat-killed, phosphorescent strain of bacteria with a living non-phosphorescent strain, you discover that some of the living cells are now phosphorescent. Which observations would provide the best evidence that the ability to fluoresce is a heritable trait? A) DNA passed from the ...
Gel Electrophoresis
... • Lithium borate (LB) is relatively new but it has low conductivity, so much higher voltage could be used, which means a shorter analysis time for routine electrophoresis. ...
... • Lithium borate (LB) is relatively new but it has low conductivity, so much higher voltage could be used, which means a shorter analysis time for routine electrophoresis. ...
hybrid DNA molecules
... replicate as a chromosomally integrated structure. This DNA may be physically isolated in intact form from either yeast or E. coli and used to transform either organism at high frequency. (iii) Vectors containing a 1.4-kilobase yeast DNA fragment that includes the centromere linked rp-l gene (YRp7) ...
... replicate as a chromosomally integrated structure. This DNA may be physically isolated in intact form from either yeast or E. coli and used to transform either organism at high frequency. (iii) Vectors containing a 1.4-kilobase yeast DNA fragment that includes the centromere linked rp-l gene (YRp7) ...
Comparison of DNA Sequences with Protein Sequences
... sequencing. The DNA sequences produced by single-pass EST sequencing and high-throughput sequencing may be of lower quality than traditional ‘‘finished’’ GenBank sequences, which are typically based on multiple sequence reads from both strands of the DNA template. As a result, EST sequences are more ...
... sequencing. The DNA sequences produced by single-pass EST sequencing and high-throughput sequencing may be of lower quality than traditional ‘‘finished’’ GenBank sequences, which are typically based on multiple sequence reads from both strands of the DNA template. As a result, EST sequences are more ...
DNA sequencing

DNA sequencing is the process of determining the precise order of nucleotides within a DNA molecule. It includes any method or technology that is used to determine the order of the four bases—adenine, guanine, cytosine, and thymine—in a strand of DNA. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.Knowledge of DNA sequences has become indispensable for basic biological research, and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in the sequencing of complete DNA sequences, or genomes of numerous types and species of life, including the human genome and other complete DNA sequences of many animal, plant, and microbial species.The first DNA sequences were obtained in the early 1970s by academic researchers using laborious methods based on two-dimensional chromatography. Following the development of fluorescence-based sequencing methods with a DNA sequencer, DNA sequencing has become easier and orders of magnitude faster.