DNA Extraction Lab - IISME Community Site
... enough so that the DNA is not broken up or shredded. In Eukaryotic cells DNA is inside the nuclear membrane, which is inside the cell membrane. In order to get the DNA out of cell both the nuclear membrane and the cell membrane must be removed. The isolated DNA can be used for DNA fingerprinting (so ...
... enough so that the DNA is not broken up or shredded. In Eukaryotic cells DNA is inside the nuclear membrane, which is inside the cell membrane. In order to get the DNA out of cell both the nuclear membrane and the cell membrane must be removed. The isolated DNA can be used for DNA fingerprinting (so ...
DNA - Duncanville ISD
... 2. Frameshift mutations: bases are inserted or deleted Are usually harmful because a mistake in DNA is carried into mRNA and results in many wrong amino acids Correct DNA: ...
... 2. Frameshift mutations: bases are inserted or deleted Are usually harmful because a mistake in DNA is carried into mRNA and results in many wrong amino acids Correct DNA: ...
Managing people in sport organisations: A strategic human resource
... Southern blot analysis for the diagnosis of fragile X syndrome. Patient DNA is simultaneously digested with restriction endonucleases EcoR1 and Eag1, blotted to a nylon membrane, and hybridized with a 32P-labeled probe adjacent to exon 1 of FMR1 (see Figure 28.1). Eag1 is a methylation-sensitive res ...
... Southern blot analysis for the diagnosis of fragile X syndrome. Patient DNA is simultaneously digested with restriction endonucleases EcoR1 and Eag1, blotted to a nylon membrane, and hybridized with a 32P-labeled probe adjacent to exon 1 of FMR1 (see Figure 28.1). Eag1 is a methylation-sensitive res ...
Chapter 10
... Nucleotide—subunit of DNA and other nucleic acids; made up of a 5-carbon sugar, a phosphate group, and a nitrogenous base ...
... Nucleotide—subunit of DNA and other nucleic acids; made up of a 5-carbon sugar, a phosphate group, and a nitrogenous base ...
Candidate gene copy number analysis by PCR and multicapillary
... Multiplex ligation-dependent probe amplification is an alternative for detection of copy number changes [16]. MLPAs probes consist of two oligonucleotides, hybridizing adjacent to each other and a sequence complementary to the target, known as the hybridization sequence. When the probes correctly hy ...
... Multiplex ligation-dependent probe amplification is an alternative for detection of copy number changes [16]. MLPAs probes consist of two oligonucleotides, hybridizing adjacent to each other and a sequence complementary to the target, known as the hybridization sequence. When the probes correctly hy ...
DNA polymerase
... 5. It can only add nucleotides to a pre-existing chain, i.e. where there is a primer – a short series of nucleotides at the 3’ end of the parental DNA to be replicated. 6. Synthesis occurs in a 5’ to 3’ direction (on the new strand) 7. The DNA nucleotides will then bind to their complementary partne ...
... 5. It can only add nucleotides to a pre-existing chain, i.e. where there is a primer – a short series of nucleotides at the 3’ end of the parental DNA to be replicated. 6. Synthesis occurs in a 5’ to 3’ direction (on the new strand) 7. The DNA nucleotides will then bind to their complementary partne ...
Chapter 6 Genes and Gene Technology Section 1 We now know
... Scientists discovered that the sequences of the bases in the DNA strand read like a book. Each set of three bases made up the “word” that coded for a specific amino acid. RECALL: Amino acids are the building blocks of proteins. So DNA tells our cells what proteins to make. Let’s find out how. The or ...
... Scientists discovered that the sequences of the bases in the DNA strand read like a book. Each set of three bases made up the “word” that coded for a specific amino acid. RECALL: Amino acids are the building blocks of proteins. So DNA tells our cells what proteins to make. Let’s find out how. The or ...
History of DNA WebQuest
... a. What did they receive the Nobel Prize for in 1962? b. What is the difference between Pauling’s structure and the actual structure of DNA? ...
... a. What did they receive the Nobel Prize for in 1962? b. What is the difference between Pauling’s structure and the actual structure of DNA? ...
Targeted Fluorescent Reporters: Additional slides
... moving DNA polymerase has a higher affinity for the correct nucleotide than an incorrect one because only the correct one can base pair with the template. 11. After nucleotide binding, but before the nucleotide is covalently bonded to the chain, the enzyme undergoes a conformational change and incor ...
... moving DNA polymerase has a higher affinity for the correct nucleotide than an incorrect one because only the correct one can base pair with the template. 11. After nucleotide binding, but before the nucleotide is covalently bonded to the chain, the enzyme undergoes a conformational change and incor ...
DNA notes
... 2. Frameshift mutations: bases are inserted or deleted Are usually harmful because a mistake in DNA is carried into mRNA and results in many wrong amino acids Correct DNA: ...
... 2. Frameshift mutations: bases are inserted or deleted Are usually harmful because a mistake in DNA is carried into mRNA and results in many wrong amino acids Correct DNA: ...
Experimental General. All the DNA manipulations and bacterial
... Together with the above mutagenic primers, in the first PCRs, BC-LIP-9F (5’CCGCCACGTACAACCAGAACTATC-3’) and PET-2R (5’-GTTATTGCTCAGCGGTGG3’) were also used, and in the second PCR, BC-LIP-9F and PET-2R were used. The conditions for the 100 µL PCR mixture were as follows: 0.5 µM each primer, 0.2 mM ea ...
... Together with the above mutagenic primers, in the first PCRs, BC-LIP-9F (5’CCGCCACGTACAACCAGAACTATC-3’) and PET-2R (5’-GTTATTGCTCAGCGGTGG3’) were also used, and in the second PCR, BC-LIP-9F and PET-2R were used. The conditions for the 100 µL PCR mixture were as follows: 0.5 µM each primer, 0.2 mM ea ...
neutral mutation
... Mutations that change protein sequences are predominantly harmful to an organism, Eg Cystic Fibrosis On occasion, the effect can be neutral or positive in a given environment. Eg. Sickle cell disease ...
... Mutations that change protein sequences are predominantly harmful to an organism, Eg Cystic Fibrosis On occasion, the effect can be neutral or positive in a given environment. Eg. Sickle cell disease ...
DNA & RNA
... * There is a person typing what is said and is creating a “court transcript”…which is really a code…shortened version…and later the transcript is translated into all the words that were said for a record. ...
... * There is a person typing what is said and is creating a “court transcript”…which is really a code…shortened version…and later the transcript is translated into all the words that were said for a record. ...
CH. 8- DNA and protein synthesis
... a. ribose, phosphate groups, and adenine b. deoxyribose, phosphate groups, and guanine c. phosphate groups, guanine, and cytosine d. phosphate groups, guanine, and thymine ____ 11. DNA is copied during a process called a. replication. b. translation. c. transcription. d. transformation. ____ 12. Whi ...
... a. ribose, phosphate groups, and adenine b. deoxyribose, phosphate groups, and guanine c. phosphate groups, guanine, and cytosine d. phosphate groups, guanine, and thymine ____ 11. DNA is copied during a process called a. replication. b. translation. c. transcription. d. transformation. ____ 12. Whi ...
Slide 1
... Deoxyribonucleic Acid (DNA) • DNA is a nucleic acid composed of units called nucleotides • Each nucleotide is made up of a sugar, a phosphate, and a nitrogen molecule • The nitrogen molecules, called bases, are: ...
... Deoxyribonucleic Acid (DNA) • DNA is a nucleic acid composed of units called nucleotides • Each nucleotide is made up of a sugar, a phosphate, and a nitrogen molecule • The nitrogen molecules, called bases, are: ...
Slide 1
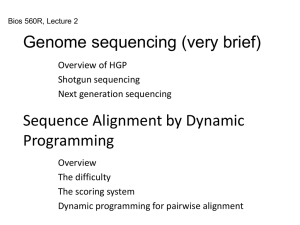
... chromosomes) is too long for sequencing directly. The DNA is randomly broken is to small pieces. The pieces are sequenced. The short sequences are assembled into long sequences based on the overlapping of fragments. ...
... chromosomes) is too long for sequencing directly. The DNA is randomly broken is to small pieces. The pieces are sequenced. The short sequences are assembled into long sequences based on the overlapping of fragments. ...
View PDF - Mvla.net
... 5. What are the main differences between DNA and RNA. DNA has deoxyribose, RNA has ribose; DNA has 2 strands, RNA has one strand; DNA has thymine, RNA has uracil. 6. Using the chart on page 303, identify the amino acids coded for by these codons: UGGCAGUGC ...
... 5. What are the main differences between DNA and RNA. DNA has deoxyribose, RNA has ribose; DNA has 2 strands, RNA has one strand; DNA has thymine, RNA has uracil. 6. Using the chart on page 303, identify the amino acids coded for by these codons: UGGCAGUGC ...
History - the Biology Department
... Cloning circumvented the limitations of traditional biochemistry, which relies on isolating molecules from a complex mixture based on their chemical idiosyncrasies. The biochemical approach is useless for purifying individual genes because chemically, they are virtually identical--each is simply a s ...
... Cloning circumvented the limitations of traditional biochemistry, which relies on isolating molecules from a complex mixture based on their chemical idiosyncrasies. The biochemical approach is useless for purifying individual genes because chemically, they are virtually identical--each is simply a s ...
Squeezing the DNA Sequences with Pattern Recognition
... (C), and thiamine (T). Interestingly the sequences of the nucleotides were exclusively bonded to be in pairs of A-T, T-A, C-G, G-C. And this discovery has opened the door to the belief that DNA was indeed capable of enough structural variety to serve as the molecule of heredity [2]. An addition to t ...
... (C), and thiamine (T). Interestingly the sequences of the nucleotides were exclusively bonded to be in pairs of A-T, T-A, C-G, G-C. And this discovery has opened the door to the belief that DNA was indeed capable of enough structural variety to serve as the molecule of heredity [2]. An addition to t ...
mb_ch13
... The Future of Genomics, continued • Microarrays – DNA microarrays, two-dimensional arrangements of cloned genes, allow researchers to compare specific genes such as those that cause cancer. ...
... The Future of Genomics, continued • Microarrays – DNA microarrays, two-dimensional arrangements of cloned genes, allow researchers to compare specific genes such as those that cause cancer. ...
幻灯片 1 - University of Texas at Austin
... the same from person to person. These sequences are called Variable Number of Tandem Repeats (VNTRs). Within the VNTRs there are sites where an enzyme can cut the DNA, and the location of these sites also varies from person to person. Cutting with the enzyme will lead to DNA fragments of differe ...
... the same from person to person. These sequences are called Variable Number of Tandem Repeats (VNTRs). Within the VNTRs there are sites where an enzyme can cut the DNA, and the location of these sites also varies from person to person. Cutting with the enzyme will lead to DNA fragments of differe ...
Microsatellite

A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from 2–5 base pairs) are repeated, typically 5-50 times. Microsatellites occur at thousands of locations in the human genome and they are notable for their high mutation rate and high diversity in the population. Microsatellites and their longer cousins, the minisatellites, together are classified as VNTR (variable number of tandem repeats) DNA. The name ""satellite"" refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying ""satellite"" layers of repetitive DNA. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists, or as simple sequence repeats (SSRs) by plant geneticists.They are widely used for DNA profiling in kinship analysis and in forensic identification. They are also used in genetic linkage analysis/marker assisted selection to locate a gene or a mutation responsible for a given trait or disease.