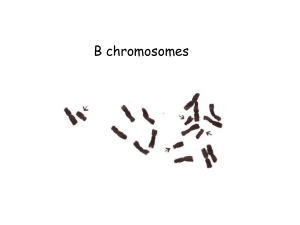chapter 7 mutation and repair of dna
... islands is that they are segments of genomic DNA at least 100 bp long with a CpG to GpC ratio of at least 0.6. These islands can be even longer and have a CpG/GpC > 0.75. They are distinctive regions of these genomes and are often found in promoters and other regulatory regions of genes. Examination ...
... islands is that they are segments of genomic DNA at least 100 bp long with a CpG to GpC ratio of at least 0.6. These islands can be even longer and have a CpG/GpC > 0.75. They are distinctive regions of these genomes and are often found in promoters and other regulatory regions of genes. Examination ...
Chpt7_RepairDNA.doc
... islands is that they are segments of genomic DNA at least 100 bp long with a CpG to GpC ratio of at least 0.6. These islands can be even longer and have a CpG/GpC > 0.75. They are distinctive regions of these genomes and are often found in promoters and other regulatory regions of genes. Examination ...
... islands is that they are segments of genomic DNA at least 100 bp long with a CpG to GpC ratio of at least 0.6. These islands can be even longer and have a CpG/GpC > 0.75. They are distinctive regions of these genomes and are often found in promoters and other regulatory regions of genes. Examination ...
The Fly Genome
... left to right starting with the X chromosome distal left arm and ending with division 100 on the distal right arm of chromosome 3. Divisions are divided into lettered subdivisions going left to right ©2000 Timothy G. Standish ...
... left to right starting with the X chromosome distal left arm and ending with division 100 on the distal right arm of chromosome 3. Divisions are divided into lettered subdivisions going left to right ©2000 Timothy G. Standish ...
Activity Name - Science4Inquiry.com
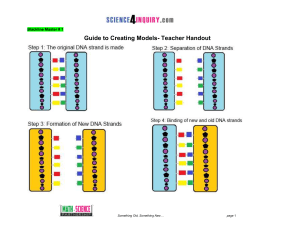
... Something Old, Something New… You are a Molecular Biologist who is studying the process of DNA replication. Your task is to build a model that represents this process. Next you will investigate where DNA replication took place in the human body and explain why it occurred. This activity is divided u ...
... Something Old, Something New… You are a Molecular Biologist who is studying the process of DNA replication. Your task is to build a model that represents this process. Next you will investigate where DNA replication took place in the human body and explain why it occurred. This activity is divided u ...
New Developments in the Embryology Laboratory
... *Mosaicism of the embryo for aneuploidy(potentially leading to false + diagnosis) *Limited no.of chromosomes studied (potentially leading to false – diagnosis) *FISH only detects the presence or the absence of the chromosomal region targeted by the probe(usually at the centromere), and does not prov ...
... *Mosaicism of the embryo for aneuploidy(potentially leading to false + diagnosis) *Limited no.of chromosomes studied (potentially leading to false – diagnosis) *FISH only detects the presence or the absence of the chromosomal region targeted by the probe(usually at the centromere), and does not prov ...
Snímek 1
... How a supernumerary B chromosome survives over time? transmission higher than Mendelian kept in populations drive (pre-meiotic, meiotic, post-meiotic) = preferential maintenance of Bs post-meiotic drive common in plants during gametophyte maturation (examples: rye, maize) ...
... How a supernumerary B chromosome survives over time? transmission higher than Mendelian kept in populations drive (pre-meiotic, meiotic, post-meiotic) = preferential maintenance of Bs post-meiotic drive common in plants during gametophyte maturation (examples: rye, maize) ...
chromosome3
... 2. Metafemale (47,XXX) a) With 3 X chromosomes (XXX), these females usually have no apparent physical abnormalities except tallness and menstrual irregularities b) Nuclei will contain two Barr bodies 3. Klinefelter syndrome (47,XXY) a) With XXY genotype, the males have underdeveloped testes, some br ...
... 2. Metafemale (47,XXX) a) With 3 X chromosomes (XXX), these females usually have no apparent physical abnormalities except tallness and menstrual irregularities b) Nuclei will contain two Barr bodies 3. Klinefelter syndrome (47,XXY) a) With XXY genotype, the males have underdeveloped testes, some br ...
Cytogenetic genotype-phenotype studies: Improving genotyping
... Deconstructing chromosomal syndromes ...
... Deconstructing chromosomal syndromes ...
hered master 4..hered 285 .. Page78
... the procedures described by Ward (1993). Great care was taken to isolate and record the origin of different embryoids as they were formed so as to eliminate any possible risk that two or more embryoids might derive from the same microspore and have the same genotype. The PGI/2 phenotype of all andro ...
... the procedures described by Ward (1993). Great care was taken to isolate and record the origin of different embryoids as they were formed so as to eliminate any possible risk that two or more embryoids might derive from the same microspore and have the same genotype. The PGI/2 phenotype of all andro ...
The nucleotide sequence of Saccharomyces cerevisiae
... telomere shows good homology with the telomeres of several other chromosomes. There are also several other internal chromosomal regions with long-range homology to other chromosomes. The largest of these is an area common to chromosomes IX and XIV, occurring at 89,233–186,363 and 478,568–616,076, re ...
... telomere shows good homology with the telomeres of several other chromosomes. There are also several other internal chromosomal regions with long-range homology to other chromosomes. The largest of these is an area common to chromosomes IX and XIV, occurring at 89,233–186,363 and 478,568–616,076, re ...
GENECLEAN® Kit
... When working with radio-labeled DNA, add 1 µl of Label Block to each 10 µl of GLASSMILK® and incubate for 5 minutes at room temperature before adding to the DNA/NaI solution in the next step. 5. Add GLASSMILK® to the DNA/NaI solution as calculated in Step 3. Vortex gently or stir with a pipet tip to ...
... When working with radio-labeled DNA, add 1 µl of Label Block to each 10 µl of GLASSMILK® and incubate for 5 minutes at room temperature before adding to the DNA/NaI solution in the next step. 5. Add GLASSMILK® to the DNA/NaI solution as calculated in Step 3. Vortex gently or stir with a pipet tip to ...
Intriguing evidence of translocations in Discus fish
... polyploidization (Thompson, 1976); it might also have arisen through chromosomal rearrangements, such as pericentric inversions, translocations and fissions/ fusions (Mesquita et al., 2008). The different karyotypic formula reported for different Symphysodon species may include errors, as the level ...
... polyploidization (Thompson, 1976); it might also have arisen through chromosomal rearrangements, such as pericentric inversions, translocations and fissions/ fusions (Mesquita et al., 2008). The different karyotypic formula reported for different Symphysodon species may include errors, as the level ...
Genetic Markers for Sex Identification in Forensic DNA Analysis
... The PCR products of AMELX and AMELY can be discriminated from one another using primers flanking a 6 bp deletion in the third intron of AMELX that is not present in AMELY [5,8-10]. The most commonly used amelogenin primer sets are those designed by Sullivan et al., producing AMELX/AMELY amplicons of ...
... The PCR products of AMELX and AMELY can be discriminated from one another using primers flanking a 6 bp deletion in the third intron of AMELX that is not present in AMELY [5,8-10]. The most commonly used amelogenin primer sets are those designed by Sullivan et al., producing AMELX/AMELY amplicons of ...
Compiling DNA strand displacement reactions using a functional
... Previous work has shown that synthetic DNA circuits can be used to implement computational systems including digital logic circuits [1], neural networks [2] and gameplaying automata [3]. In this setting, DNA is used both as an information carrier and as an engineering material, simultaneously. Furth ...
... Previous work has shown that synthetic DNA circuits can be used to implement computational systems including digital logic circuits [1], neural networks [2] and gameplaying automata [3]. In this setting, DNA is used both as an information carrier and as an engineering material, simultaneously. Furth ...
Agarose Gel Electrophoresis
... PHOTOGRAPHY OF DNA IN AGAROSE GELS DNA can be photographed in agarose gels stained with ethidium bromide by illumination with UV light (>2500 µW/cm2). A UV transilluminator is typically used for this purpose, and commercial models are available designed specifically for DNA visualization and photogr ...
... PHOTOGRAPHY OF DNA IN AGAROSE GELS DNA can be photographed in agarose gels stained with ethidium bromide by illumination with UV light (>2500 µW/cm2). A UV transilluminator is typically used for this purpose, and commercial models are available designed specifically for DNA visualization and photogr ...
Agarose Gel Electrophoresis
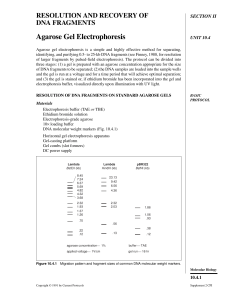
... distance judged sufficient for separation of the DNA fragments. If ethidium bromide has been incorporated into the gel, the DNA can be visualized by placing the gel on a UV light source and can be photographed directly (see support protocol). Gels run in the absence of ethidium bromide can be staine ...
... distance judged sufficient for separation of the DNA fragments. If ethidium bromide has been incorporated into the gel, the DNA can be visualized by placing the gel on a UV light source and can be photographed directly (see support protocol). Gels run in the absence of ethidium bromide can be staine ...
genotyping arabidopsis - STLCC.edu :: Users` Server
... the buffer because magnesium acts as a co-enzyme, fitting into the three dimensional structure of the protein to activate it. Too much magnesium can reduce the ability of the polymerase to stay attached to the template (i.e., fidelity is decreased), so the right concentration (1.0 – 3.0 mM) is cruci ...
... the buffer because magnesium acts as a co-enzyme, fitting into the three dimensional structure of the protein to activate it. Too much magnesium can reduce the ability of the polymerase to stay attached to the template (i.e., fidelity is decreased), so the right concentration (1.0 – 3.0 mM) is cruci ...
Initiation of recombination suppression and PAR formation during
... not generally subject to strong purifying selection, were used in this analysis. The mutation direction of each substitution was inferred in each species relative to the homolog nucleotide site in the other two species by the maximum parsimony principle. In the result, the G+C content was higher in ...
... not generally subject to strong purifying selection, were used in this analysis. The mutation direction of each substitution was inferred in each species relative to the homolog nucleotide site in the other two species by the maximum parsimony principle. In the result, the G+C content was higher in ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.