Patient with syndromic cleft lip-palate, mosaic karyotype and
... the assumption that haploinsufficiency, at least for some of the genes, in the aberrant region has a direct effect on specific development processes (6, 7). Trisomy chromosome 13 (Patau syndrome) might be the most known chromosomal abnormality which includes CLP as a common feature, but there are on ...
... the assumption that haploinsufficiency, at least for some of the genes, in the aberrant region has a direct effect on specific development processes (6, 7). Trisomy chromosome 13 (Patau syndrome) might be the most known chromosomal abnormality which includes CLP as a common feature, but there are on ...
Supplementary Notes - Word file (74 KB )
... described6 and is indicated in the schematic of Fig. 1a. In brief, an 18-mer (oligo 1: 5’-ATTCCGATAGTGACTACA-3’) was 5’-32P-labelled using T4 polynucleotide kinase (New England Biolabs) and 50 µCi [-32P]-ATP (GE Healthcare) for 15 mins at 37°C followed by 15 mins chase using 0.5 mM unlabelled ATP. ...
... described6 and is indicated in the schematic of Fig. 1a. In brief, an 18-mer (oligo 1: 5’-ATTCCGATAGTGACTACA-3’) was 5’-32P-labelled using T4 polynucleotide kinase (New England Biolabs) and 50 µCi [-32P]-ATP (GE Healthcare) for 15 mins at 37°C followed by 15 mins chase using 0.5 mM unlabelled ATP. ...
DNA Ladder, Supercoiled (D5292) - Datasheet - Sigma
... Mix an appropriate volume of the Supercoiled DNA Ladder with gel loading buffer (Product No. G2526) to the desired loading concentration. Typically 0.2 µg per well (0.02 µg/µl, 10 µl load) is sufficient to be seen using ethidium bromide staining. The recommended agarose gel concentration is 0.7% (Pr ...
... Mix an appropriate volume of the Supercoiled DNA Ladder with gel loading buffer (Product No. G2526) to the desired loading concentration. Typically 0.2 µg per well (0.02 µg/µl, 10 µl load) is sufficient to be seen using ethidium bromide staining. The recommended agarose gel concentration is 0.7% (Pr ...
Document
... • Progeny = ½ mother + ½ father DNA • Progeny = average of mother & father BVs • Assumed full sibs were identical • Available SNP information can be used to supplement the traditional approach • See difference in full-sibs at birth ...
... • Progeny = ½ mother + ½ father DNA • Progeny = average of mother & father BVs • Assumed full sibs were identical • Available SNP information can be used to supplement the traditional approach • See difference in full-sibs at birth ...
Dna rEPLICATION - Manning`s Science
... Replication begins in 2 directions from the origins as a region of DNA is unwound. Replication proceeds towards the direction of the replication fork on one strand, and away from the fork on the other. In eukaryotes, more than one replication fork may exist on a DNA molecule. A replication bubbl ...
... Replication begins in 2 directions from the origins as a region of DNA is unwound. Replication proceeds towards the direction of the replication fork on one strand, and away from the fork on the other. In eukaryotes, more than one replication fork may exist on a DNA molecule. A replication bubbl ...
DNA
... polymerase III: This need RNA primer, because DNA polymerase III can’t join the first two nucleotides to start the new strand, instead it adds the nucleotides to the existing RNA primer. RNA primer is a short segment of RNA (8-10 nucleotides with free 3' OH end) consisting of RNA bases complementary ...
... polymerase III: This need RNA primer, because DNA polymerase III can’t join the first two nucleotides to start the new strand, instead it adds the nucleotides to the existing RNA primer. RNA primer is a short segment of RNA (8-10 nucleotides with free 3' OH end) consisting of RNA bases complementary ...
Breeding - Farming Ahead
... who start recording the parentage of their sheep can take advantage of new genetic markers as they become available. The research team is using its parentage trials to measure important commercial traits such as fleece weight and fibre diameter, and subjective measurements such as colour and style o ...
... who start recording the parentage of their sheep can take advantage of new genetic markers as they become available. The research team is using its parentage trials to measure important commercial traits such as fleece weight and fibre diameter, and subjective measurements such as colour and style o ...
Sea Urchin Genome
... Steps in the Atlas Assembly System. (1) Trim off vector and low-quality portions of reads. (2) Count k-mers in WGS reads, saving the overall distribution plus specific counts for k-mers with copy number above a threshold. (3) Align BAC and WGS reads sharing rare k-mers and save overlap edges for hig ...
... Steps in the Atlas Assembly System. (1) Trim off vector and low-quality portions of reads. (2) Count k-mers in WGS reads, saving the overall distribution plus specific counts for k-mers with copy number above a threshold. (3) Align BAC and WGS reads sharing rare k-mers and save overlap edges for hig ...
Recitation Notes for RDM Day 1 1. Module Overview –
... This is a method used to separate nucleic acids on the basis of size (and shape, in some cases). DNA gels are made of agarose, a highly purified agar, heated and dissolved in a buffer solution. The agarose molecules, when cooled, form a matrix with pores between them. The more concentrated the agaro ...
... This is a method used to separate nucleic acids on the basis of size (and shape, in some cases). DNA gels are made of agarose, a highly purified agar, heated and dissolved in a buffer solution. The agarose molecules, when cooled, form a matrix with pores between them. The more concentrated the agaro ...
A Simply Fruity DNA Extraction
... All organisms are made up of cells, from simple single-‐cell bacteria to multi-‐cell humans and plants. It doesn’t matter if you are human, a bacteria or a strawberry, every cell contains deoxyribonucleic ...
... All organisms are made up of cells, from simple single-‐cell bacteria to multi-‐cell humans and plants. It doesn’t matter if you are human, a bacteria or a strawberry, every cell contains deoxyribonucleic ...
Genomics I - Faculty Web Pages
... combinatorial fluorescence and digital imaging microscopy. PNAS. 89: 4.1388-92. 1992. Courtesy Thomas Ried ...
... combinatorial fluorescence and digital imaging microscopy. PNAS. 89: 4.1388-92. 1992. Courtesy Thomas Ried ...
Casework Genetics Uses Illumina Technologies to Decipher
... Believing that such ultra-high–density SNP panels would be more effective in analyzing complex DNA mixtures—the result of co-mingled body fluids or multi-person handled evidence, such as gun grips— Dr. McElfresh and his team compared a number of platforms for suitability in forensic testing. They ch ...
... Believing that such ultra-high–density SNP panels would be more effective in analyzing complex DNA mixtures—the result of co-mingled body fluids or multi-person handled evidence, such as gun grips— Dr. McElfresh and his team compared a number of platforms for suitability in forensic testing. They ch ...
Plasmid Isolation Using Alkaline Lysis
... long strands of E. coli DNA, entangled in the remnants of lysed cells, are preferentially removed. Because each of the complementary strands of plasmid DNA is a covalently closed circle, the strands cannot be separated (without breaking one of them) by conditions such as exposure to mild alkali (up ...
... long strands of E. coli DNA, entangled in the remnants of lysed cells, are preferentially removed. Because each of the complementary strands of plasmid DNA is a covalently closed circle, the strands cannot be separated (without breaking one of them) by conditions such as exposure to mild alkali (up ...
Extraction of DNA from an Onion
... Extraction of DNA from an Onion Molecular biologists and biochemists are involved with research in finding out as much as possible about the DNA in plants and animals. Although DNA was discovered in the 1950’s, there still remains a lot to be known about it, especially how it is used to determine th ...
... Extraction of DNA from an Onion Molecular biologists and biochemists are involved with research in finding out as much as possible about the DNA in plants and animals. Although DNA was discovered in the 1950’s, there still remains a lot to be known about it, especially how it is used to determine th ...
7.1 DNA Introduction
... 2. Cytosine makes up 38% of the nucleotides in a sample of DNA from an organism. What percent of the nucleotides in this sample will be thymine? A. B. C. D. E. ...
... 2. Cytosine makes up 38% of the nucleotides in a sample of DNA from an organism. What percent of the nucleotides in this sample will be thymine? A. B. C. D. E. ...
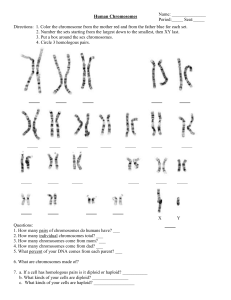
Human Chromosomes
... Directions: 1. Color the chromosome from the mother red and from the father blue for each set. 2. Number the sets starting from the largest down to the smallest, then XY last. 3. Put a box around the sex chromosomes. 4. Circle 3 homologous pairs. ...
... Directions: 1. Color the chromosome from the mother red and from the father blue for each set. 2. Number the sets starting from the largest down to the smallest, then XY last. 3. Put a box around the sex chromosomes. 4. Circle 3 homologous pairs. ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.