THE GENOMIC SEQUENCING TECHNIQUE George M. Church and
... that one normally visualizes wil l be broken up by the sequencing reactions into some several hundred fragments. Becau se we gene rally cleave at les s than 1 hit / SOO nucleotides , in such a way as to l eave lar ge amounts o f the original material unr eacted, only a few tenths of a percent of the ...
... that one normally visualizes wil l be broken up by the sequencing reactions into some several hundred fragments. Becau se we gene rally cleave at les s than 1 hit / SOO nucleotides , in such a way as to l eave lar ge amounts o f the original material unr eacted, only a few tenths of a percent of the ...
Valuing neo-native species
... as underrepresented in research and wider environmental industries. GIS skills are also highly valuable across a wide range of research areas. Analysis of palaeoecological and present-day vegetation data and modelling of tree ring data all involve complex multivariate statistical methods that will e ...
... as underrepresented in research and wider environmental industries. GIS skills are also highly valuable across a wide range of research areas. Analysis of palaeoecological and present-day vegetation data and modelling of tree ring data all involve complex multivariate statistical methods that will e ...
document
... • To identify all genes • To map all genes • To sequence all 3 billions nucleotides pairs • To create data bases • To develop sequencing methods (more fast, more efficient) • To develop new data analysis methods • To identify the ethical, legal and social problems generated by the project ...
... • To identify all genes • To map all genes • To sequence all 3 billions nucleotides pairs • To create data bases • To develop sequencing methods (more fast, more efficient) • To develop new data analysis methods • To identify the ethical, legal and social problems generated by the project ...
Biotechnology
... completely map and sequence human chromosomes (completed April 2003) • This has been important in – identifying genes that are related to disease. – Determining human relationships with other species. ...
... completely map and sequence human chromosomes (completed April 2003) • This has been important in – identifying genes that are related to disease. – Determining human relationships with other species. ...
MISCELLANEOUS NOTES 1. A Glimpse on Human Genome
... scientists to identify all of the genes contributing to a given disease state, leading to a more accurate diagnosis and precise classification of disease severity. In addition, healthy patients can know the diseases for which they are at risk, giving them the opportunity to make beneficial lifestyle ...
... scientists to identify all of the genes contributing to a given disease state, leading to a more accurate diagnosis and precise classification of disease severity. In addition, healthy patients can know the diseases for which they are at risk, giving them the opportunity to make beneficial lifestyle ...
Contemporary Biology Per
... so genetically similar, crossing individuals of the same breed increases the chances of _______ alleles pairing, which can lead to an increase in genetic ________. 14. A ___________ is an inheritable change in genetic information. Though most of the time DNA replication occurs perfectly, every once ...
... so genetically similar, crossing individuals of the same breed increases the chances of _______ alleles pairing, which can lead to an increase in genetic ________. 14. A ___________ is an inheritable change in genetic information. Though most of the time DNA replication occurs perfectly, every once ...
Metatranscriptomic analysis of the Gut microbial community
... Influence of the gut microbiome on adipogenesis. Different dietary molecules can result in changes in the gut microbial flora. The fermentation of carbohydrates and other nutrients by the gut microbiota on a high fat/high carbohydrate diet can result in an increase SCFA concentration and can result ...
... Influence of the gut microbiome on adipogenesis. Different dietary molecules can result in changes in the gut microbial flora. The fermentation of carbohydrates and other nutrients by the gut microbiota on a high fat/high carbohydrate diet can result in an increase SCFA concentration and can result ...
Slovgen s
... Genotype MDR1 +/– or N/P (carrier): Subjects with confirmed heterozygous genotype are carriers. Defective gene can be transmitted to offspring. Unwanted side effects are unlikely to occur but cannot be excluded. Genotype MDR1 –/– or P/P (affected): Particular caution is necessary in case an individu ...
... Genotype MDR1 +/– or N/P (carrier): Subjects with confirmed heterozygous genotype are carriers. Defective gene can be transmitted to offspring. Unwanted side effects are unlikely to occur but cannot be excluded. Genotype MDR1 –/– or P/P (affected): Particular caution is necessary in case an individu ...
DNA Analysis in China
... DNA Analysis in China by Hu Lan Genetics Laboratory, Institute of Forensic Sciences People’s Republic of China The Genetics Laboratory of the Institute of Forensic Sciences was the first DNA analysis unit established in China and is China’s central and main DNA profiling laboratory. The laboratory, ...
... DNA Analysis in China by Hu Lan Genetics Laboratory, Institute of Forensic Sciences People’s Republic of China The Genetics Laboratory of the Institute of Forensic Sciences was the first DNA analysis unit established in China and is China’s central and main DNA profiling laboratory. The laboratory, ...
Report Template for Positive Diagnosis Result
... 1. This assay does not detect large deletions or duplications and has limited ability to identify small insertions and deletions. This test is also has limited ability to detect mosaicism. 2. The assay does not detect variants located: 1) outside the captured exome, 2) in regions of insufficient cov ...
... 1. This assay does not detect large deletions or duplications and has limited ability to identify small insertions and deletions. This test is also has limited ability to detect mosaicism. 2. The assay does not detect variants located: 1) outside the captured exome, 2) in regions of insufficient cov ...
No Slide Title
... cyanobacteria (Synechocystis sp. PCC 6803, Synechococcus sp. WH 5701) strains were grown directly on a transparent, conductive anode (indium tin oxide-coated polyethylene terephthalate) and power generation under light and dark conditions was evaluated using a single-chamber bio-photovoltaic cell (B ...
... cyanobacteria (Synechocystis sp. PCC 6803, Synechococcus sp. WH 5701) strains were grown directly on a transparent, conductive anode (indium tin oxide-coated polyethylene terephthalate) and power generation under light and dark conditions was evaluated using a single-chamber bio-photovoltaic cell (B ...
Fish sampling - BioMed Central
... kidney, brain, white muscle and posterior intestine, were dissected out and immediately frozen in cryo tubes in liquefied nitrogen and stored at –80°C before further processing. After weight and length determination, the gender of the individuals were determined by examination of the gonads. RNA ext ...
... kidney, brain, white muscle and posterior intestine, were dissected out and immediately frozen in cryo tubes in liquefied nitrogen and stored at –80°C before further processing. After weight and length determination, the gender of the individuals were determined by examination of the gonads. RNA ext ...
Iterative literature searching
... Large negative SAM score: gene expressed more highly in Type I lesions. ...
... Large negative SAM score: gene expressed more highly in Type I lesions. ...
Package `PoissonSeq`
... of the 12 samples are rep(c(1, 2), 6). The first 80 genes are set to be overexpressed in Class 2, and the next 20 genes are set to be underexpressed in Class 2. The other 900 genes are null. This is a Poisson-distributed data. Usage data(dat) Format A list. n the count matrix y the outcome vector ty ...
... of the 12 samples are rep(c(1, 2), 6). The first 80 genes are set to be overexpressed in Class 2, and the next 20 genes are set to be underexpressed in Class 2. The other 900 genes are null. This is a Poisson-distributed data. Usage data(dat) Format A list. n the count matrix y the outcome vector ty ...
The Two Percent Difference
... It is important before discussing bioethics to understand what the two percent difference is between humans and chimpanzees. “Humans and chimps each have somewhere between 20,000 and 30,000 genes,” (Sapolsky, 2007, p. 45) thus it is likely to have differences in each gene. Sapolsky gives the interes ...
... It is important before discussing bioethics to understand what the two percent difference is between humans and chimpanzees. “Humans and chimps each have somewhere between 20,000 and 30,000 genes,” (Sapolsky, 2007, p. 45) thus it is likely to have differences in each gene. Sapolsky gives the interes ...
Lecture Notes - Course Notes
... ester bonds to its 3' to 5' hydroxyl group (i.e. phosphodiester bonds are formed between adjacent deoxyribose units). At the 1' position of the sugar ring is one of 4 nitrogen-containing bases. Two of these, Adenine and Guanine are purines and the other two, Cytosine and Thymine are pyrimidines. The ...
... ester bonds to its 3' to 5' hydroxyl group (i.e. phosphodiester bonds are formed between adjacent deoxyribose units). At the 1' position of the sugar ring is one of 4 nitrogen-containing bases. Two of these, Adenine and Guanine are purines and the other two, Cytosine and Thymine are pyrimidines. The ...
16S rRNA - Mesa Biological Indicators
... Genetic information is conserved throughout the microorganism’s life cycle. It is independent of stage of growth or even viability and is currently the most reliable resource for bacterial identification. One widely accepted genetic determination uses the 16S rRNA. This analysis uses the RNA type fo ...
... Genetic information is conserved throughout the microorganism’s life cycle. It is independent of stage of growth or even viability and is currently the most reliable resource for bacterial identification. One widely accepted genetic determination uses the 16S rRNA. This analysis uses the RNA type fo ...
Gene Set Analysis with Phenotypic Screening Data Results and Validation Purpose
... • Sensitivity analyses of the net and absolute methods have been conducted to measure the robustness of the techniques and detect false positive gene sets • The analysis was run on a viral infection cell proliferation assay then the significant sets were clustered (below). The themes are consistent ...
... • Sensitivity analyses of the net and absolute methods have been conducted to measure the robustness of the techniques and detect false positive gene sets • The analysis was run on a viral infection cell proliferation assay then the significant sets were clustered (below). The themes are consistent ...
Quiz 3 review sheet
... • Describe the conditions that, if they change, will have an impact on allele frequencies over time (Hardy Weinberg Equilibrium) • Explain how and why non-coding regions are used for DNA profiling • Interpret data from genome screening • Describe the conditions that are important for the “Hardy Wein ...
... • Describe the conditions that, if they change, will have an impact on allele frequencies over time (Hardy Weinberg Equilibrium) • Explain how and why non-coding regions are used for DNA profiling • Interpret data from genome screening • Describe the conditions that are important for the “Hardy Wein ...
Identification of disease genes Mutational analyses Monogenic
... HiSeq 2000: Up to 600 Gb per run, in 11 days 2 x 100 nt read length, two billion paired-end reads/run. In a single run, sequence two human genomes at ~30x coverage (read depth) for $3-5,000 (USD) per genome. ...
... HiSeq 2000: Up to 600 Gb per run, in 11 days 2 x 100 nt read length, two billion paired-end reads/run. In a single run, sequence two human genomes at ~30x coverage (read depth) for $3-5,000 (USD) per genome. ...
1 - Evergreen Archives
... you explain Drosophila’s faster replication of a much bigger genome? The genome of the Drosophila is organized in linear chromosomes. during DNA replication, each chromosome may have many replication bubbles/forks operating on each chromosome, speeding up the process. 7. In each of the following cas ...
... you explain Drosophila’s faster replication of a much bigger genome? The genome of the Drosophila is organized in linear chromosomes. during DNA replication, each chromosome may have many replication bubbles/forks operating on each chromosome, speeding up the process. 7. In each of the following cas ...
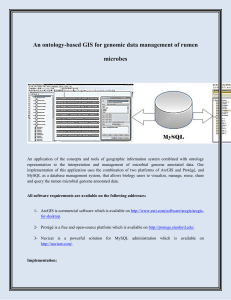
An ontology-based GIS for genomic data management of rumen
... 1- Download the ArcGIS (free trial), Protégé (free) and Navicat (free trial) 2- Install ArcGIS and then softwares Protégé and Navicat 3- Get a sample excel file containing genome annotated data http://patricbrc.org/portal/portal/patric/Home. ...
... 1- Download the ArcGIS (free trial), Protégé (free) and Navicat (free trial) 2- Install ArcGIS and then softwares Protégé and Navicat 3- Get a sample excel file containing genome annotated data http://patricbrc.org/portal/portal/patric/Home. ...
Explain the difference between the following types of genome maps
... eukaryotic genomes are much larger and have many more genes than prokaryotes, the size and complexity of an organism is not directly related to the number b r off genes it has. h F ...
... eukaryotic genomes are much larger and have many more genes than prokaryotes, the size and complexity of an organism is not directly related to the number b r off genes it has. h F ...
Identification of rare cancer driver mutations by network reconstruction
... – Network effect (linear pathway, parallel pathway) – Low sample size – Random mutation ...
... – Network effect (linear pathway, parallel pathway) – Low sample size – Random mutation ...
Introduction continued
... To obtain maps and sequences Produces nearly data that have errors (so algorithms are to be extended to handle errors. Virus and bacteria (organisms most used in genetic research) Virus consists of a protein cap (capsid) with DNA (or RNA) inside - cells starts producing-coded proteins which promotes ...
... To obtain maps and sequences Produces nearly data that have errors (so algorithms are to be extended to handle errors. Virus and bacteria (organisms most used in genetic research) Virus consists of a protein cap (capsid) with DNA (or RNA) inside - cells starts producing-coded proteins which promotes ...
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Recent studies use either ""shotgun"" or PCR directed sequencing to get largely unbiased samples of all genes from all the members of the sampled communities. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale and detail than before.