* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Leukaemia Section t(3;8)(q26;q24) Atlas of Genetics and Cytogenetics in Oncology and Haematology
Medical genetics wikipedia , lookup
Public health genomics wikipedia , lookup
Gene desert wikipedia , lookup
X-inactivation wikipedia , lookup
Minimal genome wikipedia , lookup
Non-coding RNA wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
RNA silencing wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome evolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genomic imprinting wikipedia , lookup
Oncogenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
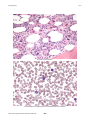
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Leukaemia Section Mini Review t(3;8)(q26;q24) Pei Lin Department of Hematopathology, Box 72, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030, USA Published in Atlas Database: November 2007 Online updated version: http://AtlasGeneticsOncology.org/Anomalies/t0308q26q24ID1463.html DOI: 10.4267/2042/38610 This work is licensed under a Creative Commons Attribution-Non-commercial-No Derivative Works 2.0 France Licence. © 2008 Atlas of Genetics and Cytogenetics in Oncology and Haematology Phenotype / cell stem origin Identity Mostly AML FAB-M2 or FAB-M-4 subtype. Etiology Unclear, may be secondary to chemotherapy. Epidemiology 10 cases reported so far in the literature, less than 1% of AML cases. Cytology Acute myeloid leukemia of mostly M2, M4 or M5 FAB subtype or high grade MDS. Marked trilineage dysplasia and megakaryocytic hyperplasia, may be associated with peripheral blood thrombocytosis giving the so-call 3q21q26 syndrome. Treatment Chemotherapy; may responds to thalidomide or arsenic better than conventional chemotherapy. Evolution Myelodysplastic syndrome progress to acute myeloid leukemia. G-banding, t(3;8)(q26;q24) Clinics and pathology Prognosis Poor. Disease Acute myeloid leukemia, de novo myelodysplastic syndrome or therapy related myelodysplastic syndrome. Atlas Genet Cytogenet Oncol Haematol. 2008;12(6) 463 t(3;8)(q26;q24) Lin P Dysplastic myeloid elements. Increased dysplastic megakaryocytes and increased blasts in the interstitium. Atlas Genet Cytogenet Oncol Haematol. 2008;12(6) 464 t(3;8)(q26;q24) Lin P is from telomere to centromere. EVI1 gene may be transcribed in different isoform which may have different oncogenic effect. Protein 1051 amino acids; 118335 Da. Nuclear location, contains 10 C2H2-type zinc fingers. Cytogenetics Note: The breakpoint on 3q26 may lie in EVI1 or MDS1 genes. The breakpoint on 8q24 is distal to the PVT1 gene, a MYC activator gene in mice. The t(3;8) is frequently associated with -7. It also can be an isolated finding. PVT1/C-MYC (pvt-1 (murine) oncogene homolog, MYC activator) Probes EVI1/MDS1 RP11-115B16, RP11-114D6; Vysis C-MYC; Breakapart distal; Probe (green). Location: 8q24 Note: The RNA function of pvt1 is unknown. References Genes involved and Proteins Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB, Verhoef G, Verdonck LF, Ossenkoppele GJ, Sonneveld P, de Greef GE, Löwenberg B, Delwel R. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood 2003;101(3):837-845. EVI1/MDS1 Location: 3q26.2 Note: Aberrant EVI1 expression usually occurs in AML, MDS or CML-BC as a result of translocation involving 3q26. The most common ones are inv(3)(q21q26), t(3;3) and t(3;21)(q26;q22). The partner genes of EVI1 are identified as Ribophorin I in inv(3)(q21q26) and t(3;3), AML/ MDS1 /EAP in t(3;21), and ETV6 in t(3;12), respectively. Others involving t(3;12), t(2;3)(p13;q26), t(3;17)(q26;q22) and t(3;13)(q26;q13-14) are uncommon. Aberrant EVI1 expression also occurs in 10% of acute myeloid leukemia without involving 3q26 and is also correlated with an adverse outcome. DNA / RNA 16 exons spanning 64.2 Kb. Transcriptional orientation Atlas Genet Cytogenet Oncol Haematol. 2008;12(6) Poppe B, Dastugue N, Vandesompele J, Cauwelier B, De Smet B, Yigit N, De Paepe A, Cervera J, Recher C, De Mas V, Hagemeijer A, Speleman F. EVI1 is consistently expressed as principal transcript in common and rare recurrent 3q26 rearrangements. Genes Chromosomes Cancer 2006;45(4):349-356. Lennon PA, Abruzzo LV, Medeiros LJ, Cromwell C, Zhang X, Yin CC, Kornblau SM, Konopieva M, Lin P. Aberrant EVI1 expression in acute myeloid leukemias associated with the t(3;8)(q26;q24). Cancer Genet Cytogenet 2007;177(1):37-42. This article should be referenced as such: Lin P. t(3;8)(q26;q24). Atlas Genet Haematol.2008;12(6):463-465. 465 Cytogenet Oncol