* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Effects of adenovirus delivered Flt
Epigenetics in stem-cell differentiation wikipedia , lookup
DNA vaccination wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Genome evolution wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Point mutation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Oncogenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Genome (book) wikipedia , lookup
Protein moonlighting wikipedia , lookup
Minimal genome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Genomic imprinting wikipedia , lookup
Designer baby wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
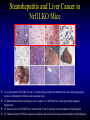
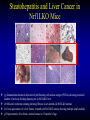
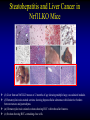
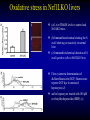
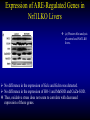
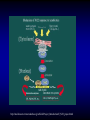
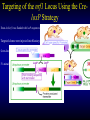
Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia Zhenrong Xu, Linyun Chen, Laura Leung, T. S. Benedict Yen, Candy Lee, and Jefferson Y. Chan. Proc Natl Acad Sci U S A. 2005 March 15; 102(11): 4120–4125. Introduction The term oxidative stress encompasses a broad spectrum of circumstances that cause a change in the cellular redox status such as an increased production of free radical species within the cell or by pro-oxidant xenobiotics that are thiol reactive and mimic an oxidative insult. The antioxidant response element (ARE) is a cis-acting enhancer sequence that mediates transcriptional activation of genes in cells exposed to oxidative stress In keeping with this, genes that are regulated by the ARE encode proteins that help control the cellular redox status and defend the cell against oxidative damage Proteins that are encoded by the ARE gene battery include enzymes associated with glutathione biosynthesis, redox proteins with active sulfhydryl moieties, and drug-metabolizing enzymes ARE Annual Review of Pharmacology and Toxicology. 43: 233-260 Sequences of ARE Found in Different Genes The consensus sequence for ARE is 5’-TGACnnnGC-3’ Annual Review of Pharmacology and Toxicology. 43: 233-260 Regulation of ARE Regulation of ARE function is mediated by various basic leucine zipper (bZIP) transcription factors including members of the ‘‘cap n collar’’ (CNC)-bZIP, such as Nrf1 and Nrf2, and small-Maf family of proteins. 51qe.cn/pic/30/ 11/18/07707.jpg Nrf1 and Nrf2 Nrf1 and Nrf2 are CNC-bZIP proteins, and they function as obligate heterodimers by complexing with small-Maf and other bZIP proteins. An important role for Nrf2 in xenobiotic metabolism and oxidative stress response has been identified through knockout studies in mice indicating that Nrf2 is an important activator of AREs. Analysis of nrf1 and nrf1::nrf2 mutant cells suggests that Nrf1 is also involved in the oxidative stress response. The function of Nrf1 is not fully understood. Mice deficient in Nrf1 function die during development. Nrf1 and Liver Chimeric mice generated with Nrf1-deficient embryonic stem cells showed widespread apoptosis in fetal livers at late gestation, demonstrating a cell autonomous role of Nrf1 in the survival of hepatocytes. This finding suggests that Nrf1 is required for normal function of hepatocytes. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia Zhenrong Xu, Linyun Chen, Laura Leung, T. S. Benedict Yen, Candy Lee, and Jefferson Y. Chan. Proc Natl Acad Sci U S A. 2005 March 15; 102(11): 4120–4125. Hypothesis Nrf1 is critical to the oxidative stress response in the adult liver, and it plays an important role in oxidative stress-induced liver disease. Approach To bypass embryonic lethality, a Cre-lox system was used to investigate the function of Nrf1 in adult liver Targeting DNA with the Cre/lox System http://www.bioteach.ubc.ca/MolecularBiology/TargetingYourDNAWithTheCreloxSystem/ Targeting DNA with the Cre/lox System http://www.bioteach.ubc.ca/MolecularBiology/TargetingYourDNAWithTheCreloxSystem/ Tissue (Liver)-Specific Knockout Transactivator Floxed Responder X Conventional Transgenic ES-derived mouse 12 kb Albumin Promoter & enhancer Cre Floxed allele: “normal” expression Cre expression in liver Liver Liver-specific gene deletion Elsewhere “Normal” expression http://med.umich.edu/hg/EDUCATION/COURSES/HG541/Camper3.ppt#337,15,Tissue-Specific Knockou Targeting of the nrf1 Locus Using the CreloxP Strategy (a Top) Map of the targeting construct containing loxP elements flanking the terminal exon of nrf1 and the positive (PGKNEO) selection cassette, IRES-GFP cassette and a negative (DT) selection cassette (Middle) The terminal portion of the wildtype nrf1 gene, and the 5′ external probe used to detect targeted clones. (Bottom) The targeted allele. The predicted sizes of NsiI fragments of wild-type (14-kb) and targeted allele (11.5-kb) are shown. (b) Southern blot analysis of ES clones digested with NsiI and probed with the 5′ external probe. Targeting of the nrf1 locus, contd. (c Top) The nrf1-neo knockout allele. (Middle) The floxed nrf1 allele. (Bottom) The Cre-mediated recombined allele. (Right) Southern blot analysis of liver DNA obtained from control and Nrf1LKO mice. Top band: the 4kb EcoRI fragment of both the nrf1-neo and floxed nrf1 alleles detected by the indicated probe. Lower band: the 2.1kb EcoRI fragment of the recombined allele. (d) Analysis of recombined and nonrecombined floxed nrf1 allele in liver DNA. Top band: the 550-bp product amplified from the recombined floxed nrf1 allele. Bottom band: the 250-bp product amplified from the nonrecombined floxed nrf1 allele. Conclusion Nrf1LKO Mice were generated. No increase in mortality or morbidity was observed early on in Nrf1LKO mice compared with littermate controls (nrf1flox/– and AlbCre;nrf1+/–). Steatohepatitis and Liver Cancer in Nrf1LKO Mice (a) Serum concentration of alanine aminotransferase (ALT) in control and Nrf1LKO mice at 4 weeks of age. (b) Liver triglyceride levels in control and Nrf1LKO mice at 6–8 weeks of age. Steatohepatitis and Liver Cancer in Nrf1LKO Mice (c) A representative Nrf1LKO liver at 4–8 weeks of age stained with hematoxylin/eosin showing apoptosis, necrosis, inflammatory infiltrate, and vacuolated cells. (d) Immunohistochemical staining for active caspase-3 in Nrf1LKO liver showing multiple apoptotic hepatocytes. (e) Frozen section of Nrf1LKO liver stained with oil red O showing increased number of lipid droplets. (f) Cultured primary Nrf1LKO hepatocytes stained with oil red O showing increased number of lipid droplets. Steatohepatitis and Liver Cancer in Nrf1LKO Mice (g) Immunohistochemical detection of proliferating cell nuclear antigen (PCNA) showing increased number of actively dividing hepatocytes in Nrf1LKO liver. (h) Masson's trichrome staining showing fibrosis in a 6-month-old Nrf1LKO animal. (i) Gross appearance of a liver from a 4-month-old Nrf1LKO mouse showing multiple small nodules. (j) Representative liver from a control mouse at 12 months of age. Steatohepatitis and Liver Cancer in Nrf1LKO Mice (k) Liver from an Nrf1LKO mouse at 12 months of age showing multiple large, vascularized nodules. (l) Hematoxylin/eosin-stained sections showing hepatocellular adenomas with distinctive borders between tumors and parenchyma. (m) Hematoxylin/eosin stained sections showing HCC with trabecullar features. (n) Section showing HCC-containing clear cells. Conclusion Loss of Nrf1 results in steatohepatitis and spontaneous liver cancer. Question Whether oxidative stress was present before development of tumors? Oxidative stress in Nrf1LKO livers (a) Liver TBARS levels in control and Nrf1LKO mice. (b) Immunohistochemical staining for 8oxoG showing no reactivity in normal liver. (c) Immunohistochemical detection of 8oxoG-positive cells in Nrf1LKO liver. Flow cytometric determination of dichlorofluorescein (DCF) fluorescence reporter DCF dye in untreated hepatocytes (d) and in hepatocytes treated with 100 μM tert-butylhydroperoxide (tBHP) (e). Conclusion Loss of Nrf1 results in oxidative stress in hepatocytes, and oxidative injury is present in Nrf1LKO livers before tumor formation. Mutant hepatocytes also showed a severe impairment in handling additional oxidant stress. Nrf1 has been shown to regulate expression of both the catalytic and regulatory subunits of glutamyl-cysteine ligase (Gclc and Gclm). Question Whether oxidative stress resulted from altered regulation of these genes (Gclc and Gclm). Expression of ARE-Regulated Genes in Nrf1LKO Livers (a) Western blot analysis of control and Nrf1LKO livers. No difference in the expression of Gclc and Gclm was detected. No difference in the expression of HO-1 and MnSOD and CuZn-SOD. Thus, oxidative stress does not seem to correlate with decreased expression of these genes. Question Whether expression of other known ARE-dependent genes was altered in Nrf1LKO livers? PCR Primers Expression of ARE-Regulated Genes in Nrf1LKO Livers (b) RT-PCR analysis of mRNA encoding various ARE-dependent genes and lipid metabolism. 18s levels were used as control. No significant change in GSTA3, GSTA4, GSTM1, GSTM2, GSTM4, and GSTT1. Transcripts for GSTM3, GSTM6, and GSTP2 were clearly reduced in Nrf1LKO samples GSTA1 and metallothionein-1 and -2 were elevated in Nrf1LKO livers, indicating that other counteractive mechanisms against oxidative stress were induced. A subset of ARE-containing genes was affected in Nrf1LKO livers. Fatty acid metabolism by peroxisomal and microsomal pathways generates a significant source of ROS. The fatty liver phenotype in Nrf1LKO mice. Question whether these pathways (peroxisomal and microsomal pathways ) were induced in Nrf1LKO mice. Microsome Proliferation and Induction of ω-fatty Acid Oxidation in Nrf1LKO Livers (a) RT-PCR analysis of mRNA encoding various enzymes associated with β- and ω-fatty acid oxidation. PPAR-α, peroxisome proliferatoractivated receptor α; Acox1, acyl-CoA oxidase; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; Acaa1, acetyl-CoA acyltransferase 1 No increase in the PPAR-α expression in Nrf1LKO. PPAR-α did not seem to be transcriptionally activated because expression of its target genes, including Acox1, Ehhadh, and Acaa was not altered. A significant induction of cytochrome P450 4a10 and 4a14 (CYP4a10 and CYP4a14) Microsome Proliferation and Induction of ω-fatty Acid Oxidation in Nrf1LKO Livers Representative electron micrograph of control (b) and Nrf1LKO (c) livers at 2 months. Note lipid droplets, smaller and darker mitochondria, and proliferation of smooth ER in the Nrf1LKO liver. Nrf1LKO livers showed proliferation of smooth endoplasmic reticulum. Conclusion β-oxidation pathway does not seem to be induced in Nrf1LKO livers. Nrf1LKO livers showed proliferation of smooth endoplasmic reticulum. ω-oxidation of fatty acids by means of microsomal CYP4A enzymes participates in the generation of oxidative stress in Nrf1LKO livers. Summary and Conclusions No increase in mortality or morbidity was observed early on in Nrf1LKO mice compared with littermate controls (nrf1flox/– and AlbCre;nrf1+/–). Loss of Nrf1 results in steatohepatitis and spontaneous development of liver cancer. loss of Nrf1 results in oxidative stress in hepatocytes, and oxidative injury is present in Nrf1LKO livers before tumor formation. A subset of ARE-containing genes was affected in Nrf1LKO livers. β-oxidation pathway does not seem to be induced in Nrf1LKO livers. Summary and Conclusions Nrf1LKO livers showed proliferation of smooth endoplasmic reticulum. ω-oxidation of fatty acids by means of microsomal CYP4A enzymes participates in the generation of oxidative stress in Nrf1LKO livers. In Nrf1LKO livers, reactive oxygen species generated from CYP4Amediated fatty acid oxidation work synergistically with diminished expression of ARE-responsive genes to cause oxidative stress in mutant hepatocytes. The changes observed in Nrf1LKO livers characterized by the initial presence of cell death, proliferation, and inflammation, followed by the appearance of dysplastic cells and ultimately cancer, closely mimic the salient features of NASH. These Nrf1LKO animals may prove useful as a model to study the mechanisms in the development and progression of NASH and liver cancer. Critique The article is well written. Results are consistent with conclusions First report that provides genetic evidence identifying a critical role for Nrf1 attenuating oxidative stress and neoplastic growth in livers of adult mice. “We cannot rule out that Nrf1 may function as a tumor suppressor in hepatocytes, and it is also possible that Nrf1 may accelerate disease progression by regulating yet to be identified pathways.” Other ARE-containing target genes that are involved in the oxidative stress response (e.g. GSTA2 and NQO1) may be also affected by loss of Nrf1 in the liver. Critique Some discrepancies in text and figures. “Wild-type and targeted alleles yield a 14-kb and a 16-kb fragment, respectively” (Fig. 1b). Lack of controls for Fig. 2 Future Directions Identify other ARE-containing target genes that are involved in the oxidative stress response. They may be also affected by loss of Nrf1 in the liver. Determine whether the level of expression of Nrf2 changes in Nrf1LKO livers. it is possible that Nrf2 may functionally compensate for the loss of Nrf1, because several of the ARE-regulated genes examined are also known targets of Nrf2 Examine the role of other bZIP proteins and small-Maf proteins which form heterodimers with Nrf1 in the oxidative stress response. Extend this study to other tissues like kidney, lung, ..etc Thank You http://web.indstate.edu/thcme/mwking/glutathione.gif http://www.med.unibs.it/~marchesi/glu tathione.gif http://zoology.muohio.edu/oris/ZOO462/notes/images/06c_46204.jpg Omega Oxidation While the main route of fatty acid metabolism is through beta-oxidation, some minor metabolic pathways such as omega oxidation also contribute to the metabolism of fatty acids and other molecules. Omega oxidation occurs in the endoplasmic reticulum rather than the mitochondria, the site of beta-oxidation. The omega carbon in a fatty acid is the carbon furthest in the alkyl chain from the carboxylic acid. In the omega oxidation pathway, this carbon is progressively oxidized first to an alcohol and then to a carboxylic acid, creating a molecule with a carboxylic acid on both ends. The first step in the pathway is catalyzed by a cytochrome P450 mixed function oxidase and requires both oxygen and NADPH. Oxidation of the alcohol is catalyzed by an alcohol dehydrogenase and aldehyde dehydrogenase catalyzes the formation of the dicarboxylic acid. • • REGULATORY MECHANISMS CONTROLLING GENE EXPRESSION MEDIATED BY THE ANTIOXIDANT RESPONSE ELEMENT Truyen Nguyen, Philip J. Sherratt, and Cecil B. Pickett Schering-Plough Research Institute, Kenilworth, New Jersey 07033; email: [email protected] [email protected] [email protected] • The expression of genes encoding antioxidative and Phase II detoxification enzymes is induced in cells exposed to electrophilic compounds and phenolic antioxidants. Induction of these enzymes is regulated at the transcriptional level and is mediated by a specific enhancer, the antioxidant response element or ARE, found in the promoter of the enzyme's gene. The transcription factor Nrf2 has been implicated as the central protein that interacts with the ARE to activate gene transcription constitutively or in response to an oxidative stress signal. This review focuses on the molecular mechanisms whereby the trancriptional activation mediated by the interaction between the ARE and NF-E2-related factor 2 (Nrf2) is regulated. Recent studies suggest that the sequence context of the ARE, the nature of the chemical inducers, and the cell type are important for determining the activity of the enhancer in a particular gene. http://mol-devserver.tara.tsukuba.ac.jp/official/Project_Introduction/E_Nrf2_project.html http://mol-devserver.tara.tsukuba.ac.jp/official/Project_Introduction/E_Nrf2_project.html dir.niehs.nih.gov/ dirlrb/eg/proj-nrf2.htm Three major signaling pathways have been implicated in the regulation of the AREmediated transcriptional response to chemical stress. In vitro data suggest that direct phosphorylation by PKC may promote Nrf2 nuclear translocation as a mechanism leading to transcriptional activation of the ARE. Nrf2 has been proposed to be retained in the cytoplasm through an interaction with Keap1 and it is possible that phosphorylation of Nrf2 may also cause the disruption of this interaction. As a bZIP protein, Nrf2 binds to the ARE as a dimer. Although small Maf proteins have been proposed to represent the dimerizing partners for Nrf2 in the activation complex, this has not been conclusively demonstrated. The molecular mechanisms controlling the ARE-mediated transcription by the MAP kinase and PI3 kinase pathways remain to be determined Introduction The antioxidant response element (ARE) is a cis-acting enhancer sequence that mediates transcriptional activation of genes in cells exposed to oxidative stress The term oxidative stress encompasses a broad spectrum of circumstances that cause a change in the cellular redox status such as an increased production of free radical species within the cell or by pro-oxidant xenobiotics that are thiol reactive and mimic an oxidative insult. In keeping with this, genes that are regulated by the ARE encode proteins that help control the cellular redox status and defend the cell against oxidative damage Proteins that are encoded by the ARE gene battery include enzymes associated with glutathione biosynthesis, redox proteins with active sulfhydryl moieties, and drug-metabolizing enzymes Introduction The expression of genes encoding antioxidative and Phase II detoxification enzymes is induced in cells exposed to electrophilic compounds and phenolic antioxidants. Induction of these enzymes is regulated at the transcriptional level and is mediated by a specific enhancer, the antioxidant response element or ARE, found in the promoter of the enzyme's gene. The transcription factor Nrf2 (NF-E2-related factor 2) has been implicated as the central protein that interacts with the ARE to activate gene transcription constitutively or in response to an oxidative stress signal. Introduction NF-E2 is a dimeric protein originally identified as involved in the regulation of globin gene expression in hematopoietic cells. The NF-E2 protein activates gene transcription following binding to its consensus DNA binding motif 5'-TGCTGAGTCAC-3' as a heterodimer consisting of a 45-Kda and an 18-Kda subunit. The p45 subunit is a bZIP protein consisting of a transactivation domain within the N-terminal region and a basic DNA binding region/leucine zipper structure in the C-terminal region of the protein. The p18 subunit was subsequently identified as a member of the small Maf proteins containing the basic region/leucine zipper but lacking any apparent transactivation domain Introduction Whereas expression of the p45 NF-E2 subunit is restricted to hematopoietic tissues, two other related bZIP family members, Nrf1 and Nrf2, are ubiquitously expressed in a wide range of tissues and cell types. The tissue distribution profile of these proteins combined with their DNA binding motif being similar in comparison to that of the ARE led to the suggestion that ARE-responsive genes may be regulated by Nrf1 and/or Nrf2. Nrf2 was subsequently demonstrated to be involved in the transcriptional activation of other ARE-responsive genes including those encoding the human γ-GCSh and γ-GCSl subunits, mouse HO1, and rat NQO1 and GSTA2 subunits. Introduction Nrf2 is a critical protein in regulating the expression of the Gsta1 and Nqo1 genes. These data suggest that Nrf2 mediates both the basal and inducible activity of the ARE. The loss of Nrf2 resulted in a profound reduction in the expression and enzyme activities of NQO1, certain GST isoenzymes, and the γ-GCSh subunit. Current data from in vitro DNA binding assays and transfection experiments point to Nrf2 as the most important protein involved in stimulating ARE-driven transcription. The reduced expression of Phase II drug-metabolizing enzymes in Nrf2 (−/−) mice confers a sensitive phenotype to the toxic effects of carcinogens and inflammatory drugs compared to wild-type animals. Introduction Ectopic expression of Nrf1 activates the human NQO1 ARE reporter gene in HepG2 cells. Nrf1 also activates a reporter gene linked to the promoter of the γ-GCSh subunit gene and increases the intracellular level of GSH. Although the loss of Nrf1 is lethal in early embryonic development, an analysis of mouse fibroblasts derived from Nrf1 (−/−) mouse embryonic tissue showed reduced levels of GSH and of γ-GCSl subunit expression compared to wild-type cells. Interestingly, induction of this enzyme was not affected by the loss of Nrf1, as treatment of the Nrf1-deficient fibroblasts with paraquat resulted in an increased level of its mRNA. These data and those obtained from studies involving Nrf2 (−/−) mice suggest that the expression of γ-GCS genes is mediated by both Nrf1 and Nrf2, with Nrf1 being involved in basal expression and Nrf2 in the inducible expression. Introduction Current data from in vitro DNA binding assays and transfection experiments point to Nrf2 as the most important protein involved in stimulating ARE-driven transcription. The reduced expression of Phase II drug-metabolizing enzymes in Nrf2 (−/−) mice confers a sensitive phenotype to the toxic effects of carcinogens and inflammatory drugs compared to wild-type animals. Introduction Ectopic expression of Nrf1 activates the human NQO1 ARE reporter gene in HepG2 cells. Nrf1 also activates a reporter gene linked to the promoter of the γ-GCSh subunit gene and increases the intracellular level of GSH. Although the loss of Nrf1 is lethal in early embryonic development, an analysis of mouse fibroblasts derived from Nrf1 (−/−) mouse embryonic tissue showed reduced levels of GSH and of γ-GCSl subunit expression compared to wild-type cells. Interestingly, induction of this enzyme was not affected by the loss of Nrf1, as treatment of the Nrf1-deficient fibroblasts with paraquat resulted in an increased level of its mRNA. These data and those obtained from studies involving Nrf2 (−/−) mice suggest that the expression of γ-GCS genes is mediated by both Nrf1 and Nrf2, with Nrf1 being involved in basal expression and Nrf2 in the inducible expression. Overview • Transcription of many cytoprotective genes and phase II xenobiotic metabolizing genes is regulated through cis-active sequences known as antioxidant response elements (ARE). • Regulation of ARE function is mediated by various basic leucine zipper (bZIP) transcription factors including members of the ‘‘cap n collar’’ (CNC)-bZIP and small-Maf family of proteins. • Nrf1 and Nrf2 are CNC-bZIP proteins, and they function as obligate heterodimers by complexing with small-Maf and other bZIP proteins. • An important role for Nrf2 in xenobiotic metabolism and oxidative stress response had been identified through knockout studies in mice indicating that Nrf2 is an important activator of AREs. Targeting of the nrf1 Locus Using the CreloxP Strategy Exon-4 of nrf1 was flanked with loxP sequences Targeted clones were injected into blastocyst Germ-line transmission was achieved F1 animals were obtained