* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lesson08Phylogenetics
Extrachromosomal DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Molecular cloning wikipedia , lookup
Genomic library wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Non-coding DNA wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Minimal genome wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Maximum parsimony (phylogenetics) wikipedia , lookup
History of genetic engineering wikipedia , lookup
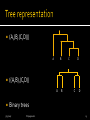
Doug Raiford Lesson 8 5/23/2017 Phylogenetics 1 Definition study of evolutionary relatedness among various groups of organisms All life evolved from a single-celled organism Cenancestor Traditional techniques: Relied upon phenotypical (observable characteristic or trait of an organism) differences 5/23/2017 Phylogenetics 2 Understanding of the origins of life Who we are? From whence did we come? If can understand biological systems and how they came to be… Why study phylogenetics? Perhaps can predict ▪ Reaction to changing environments ▪ Reaction to drugs (similar organisms will probably react similarly) What our future holds How we will evolve 5/23/2017 Phylogenetics 3 DNA similar in related organisms How would we go about measuring DNA similarity? Must align first Must use homologous genes/proteins Just count thedo locations Where genes and proteins where different come into the picture? 5/23/2017 Phylogenetics 4 Different rates of evolutionary change Organisms: different environmental factors Proteins: under different selective pressures Regions of proteins: ▪ Interiors, tightly packed, hydrophobic ▪ External loops, less important for structural integrity 5/23/2017 Phylogenetics 5 Gap of virtually any length could happen in single event Buchnera/1-356 Lactobacillus/1-363 Geobacter/1-338 Actinobacillus/1-376 Salmonella/1-353 5/23/2017 MENL----------------DKKKALDRVIMEIEKAYGKGAIMKLG-EMA MAKD----------------EKKAALDAALKKIEKNFGKGAVMRMG-EKA MTQ-----------------EREKAIELALSQIEKQFGKGAIMRLGADEA MAADNKKAQKNTVTKQIDPEQKEKALAAALAQIEKQFGKGSIMRLG-DTQ MAID---------------ENKQKALAAALGQIEKQFGKGSIMRLG-EDR Phylogenetics 6 DNA material doesn’t stay put Bacteria reproduce asexually But plasmids… Also, viruses… Finally, take-up… Not surprising… Meiosis, mitosis, translocating genes 5/23/2017 Phylogenetics 7 Horizontal gene transfer (HGT) More of a bush (especially at the base) 5/23/2017 Phylogenetics 8 Need a gene that is in all organisms (ubiquitous) The gene should be evolutionarily stable (very similar in all organisms) Should compare regions of the gene that are highly conserved 5/23/2017 Phylogenetics 9 Circular DNA found in organelles outside the nucleus No crossover: passed down in the egg Exact copy from female parent Mitochondria are the powerhouse of the cell Break-down food, release energy 5/23/2017 Phylogenetics 10 Dominant molecule in microbial ribosomes (translation) Ubiquitous, same role in every organism Highly conserved 5/23/2017 Phylogenetics 11 Multiple sequence alignments ClustalW Sound Easily familiar? identifyWhat regions that arealready highlyhave? tools do we conserved atgccgca-actgccgcaggagatcaggactttcatgaatatcatcatgcgtggga-ttcag acctccatacgtgccccaggagatctggactttcacc---tggatcatgcgaccgtacctac t-atgg-t-cgtgccgcaggagatcaggactttca-gt--g-aatcatctgg-cgc--c-aa t--tcgt-ac-tgccccaggagatctggactttcaaa---ca-atcatgcgcc-g-tc-tat aattccgtacgtgccgcaggagatcaggactttcag-t--a-tatcatctgtc-ggc--tag 5/23/2017 Phylogenetics 12 Used 16S to investigate tree of life Discovered three domains (not two) 5/23/2017 Phylogenetics 13 Build cladogram or phylogram: A tree diagram used to illustrate phylogenetic relationships Length of each branch = number of sequence changes that occurred (except in cladogram where length not used) The amount of time usually not known Sometimes utilize a dendrogram 5/23/2017 Phylogenetics 14 Each sequence taxon (plural: taxa) Each subtree = clade (long “a”) Tree length Sum of all the branch lengths Sometimes utilize a dendrogram 5/23/2017 Phylogenetics 15 Assumption of a uniform rate of mutation in the tree branches Is this reasonable? Allows some conclusions to be drawn 5/23/2017 Phylogenetics 16 Distance Maximum parsimony Maximum likelihood Main packages PAUP PHYLIP 5/23/2017 Phylogenetics 17 Unweighted pair group method with arithmetic mean (UPGMA) One of the first and simplest distance methods Basically hierarchical clustering 5/23/2017 Phylogenetics 18 Need all pair-wise distances Count up the number of columns How measure distance? where there are and Want to measure thedifferences number of mutations divide by thethe length of the that occurred since species split sequences: the probability of a mutation at a given location Distance between organism A and B is 4 2 Organism A 5/23/2017 Phylogenetics 2 Organism B 19 A A B C D 0 6 6 6 0 4 4 0 2 B All pair-wise distances C D 0 1 1 3 2 A 5/23/2017 Phylogenetics 1 B C 1 D 20 Find closest Place next to each other in tree Find average distance to rest A A B C D 0 6 6 6 0 4 4 0 2 B C D 0 1 A B CD 5/23/2017 A B CD 0 6 6 0 4 1 3 2 A 1 B C 1 D 0 Phylogenetics 21 Find the next closest Place next to each other in tree A A B CD 0 6 6 0 4 B CD 0 1 A BCD A BCD 0 6 2 0 A 5/23/2017 Phylogenetics 1 3 1 B C 1 D 22 A A B C D 0 6 6 7 0 4 5 0 3 B New distance matrix C D A 5/23/2017 Phylogenetics 0 B C D 23 C and D still closest So start tree with these two Find average distance to rest A B CD 5/23/2017 A B CD 0 6 6.5 0 4.5 A A B C D 0 6 6 7 0 4 5 0 3 B C D A 0 B C D 0 Phylogenetics 24 (A,(B,(C,D))) A B D ((A,B),(C,D)) A C B C D Binary trees 5/23/2017 Phylogenetics 25 Find the next closest Place next to each other in tree (collapse B with CD) A BCD 5/23/2017 A BCD 0 6.25 A B A B CD 0 6 6.5 0 4.5 CD A B 0 C D 0 Phylogenetics 26 5/23/2017 Phylogenetics 27