* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Eukaryotic Gene Expression
Gene nomenclature wikipedia , lookup
X-inactivation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Ridge (biology) wikipedia , lookup
Transposable element wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
RNA interference wikipedia , lookup
Polyadenylation wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
History of RNA biology wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Messenger RNA wikipedia , lookup
RNA silencing wikipedia , lookup
Designer baby wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Transcription factor wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epitranscriptome wikipedia , lookup
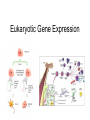
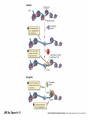
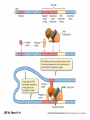
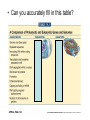
Eukaryotic Gene Expression Introduction • Every cell in a multi-cellular eukaryote does not express all its genes, all the time (usually only 3-5%) – Long-term control of gene expression in tissue = differentiation • How to prevent expression? – Regulation at transcription – Regulation after transcription Chromatin Regulation • Chromatin remodeling allows transcription – Chromatin = DNA + proteins – Chromatin coiled around histones = nucleosomes – Allows DNA to be packed into nucleus, but also physically regulates expression by making regions ‘available’ or not Chromatin Regulation Con’t • Chromatin regulation can be small-scale (gene) or large scale (chromosome) – Non-expressed = heterochromatin (condensed) – Expressed = euchromatin (relaxed) • Example of whole-chromosome regulation: Barr bodies Gene Amplification • To increase gene expression, make temporary copies of a gene, aka gene amplification • Can happen in certain tissues or stages of development – Embryos require massive volumes of rRNA (to make ribosomes); in early development there are a million+ extra rRNA genes (not able to replicate) present in nucleus Transcription Regulation • What we know from prokaryotes: – Several related genes can be transcribed together (ie. lac operon) – Need RNA Polymerase to recognize a promoter region • Why eukaryotes are different: – Genes are nearly always transcribed individually – 3 RNA Polymerases occur, requiring multiple proteins to initiate transcription Transcription Regulation Con’t • Typical prokaryotic promoter: recognition sequence + TATA box -> RNA Polymerase attachment -> transcription • Typical eukaryotic promoter: recognition sequence + TATA box + transcription factors -> RNA Polymerase II attachment -> transcription Transcription Regulation Con’t • RNA polymerase interacts w/promoter, regulator sequences, & enhancer sequences to begin transcription – Regulator proteins bind to regulator sequences to activate transcription • Found prior to promoter – Enhancer sequences bind activator proteins • Typically far from the gene • Silencer sequences stop transcription if they bind with repressor proteins Transcription Regulation Con’t • If eukaryotic genes are typically ‘alone’, how to regulate expression of several? • Conserve regulatory sequences! Now, Can You: • Explain why gene expression control is necessary in a eukaryotic cell? • Describe how expression is regulated in before & during transcription? • Tell me what differentiation is? Euchromatin? A silencer sequence? • Explain how gene expression regulation is different in eukaryotes/prokaryotes? Post-Transcription Regulation • Have mRNA variation – Alternative splicing: shuffling exons – Allows various proteins to be produced in different tissues from the same gene • Change the lifespan of mRNA – Produce micro RNA that will damage mRNA, preventing translation • Edit RNA & change the polypeptide produced – Insert or alter the genetic code Translation Regulation • mRNA present in cytosol does not necessarily get translated into proteins – Control the rate of translation to regulate gene expression • How? – Modify the 5’ cap – Feedback regulation (build up of products = less translation) Translation Regulation Con’t • Modify the lifespan of proteins: – Attach ubiquitin = target for breakdown via proteasome (woodchipper) So… • What are the ways that a cell can regulate gene expression AFTER transcription? • How can the process of RNA splicing allow one pre-mRNA to produce 5 different proteins in 5 different tissues? • And… • Can you accurately fill in this table?