* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Document
Survey
Document related concepts
Protein adsorption wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene prediction wikipedia , lookup
Chemical biology wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Ligand binding assay wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding RNA wikipedia , lookup
Expression vector wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Gene regulatory network wikipedia , lookup
Transcript
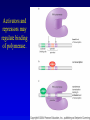
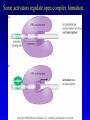
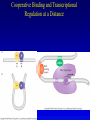
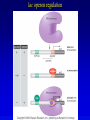
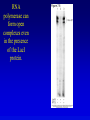
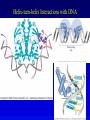
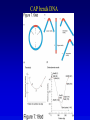
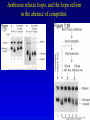
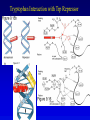
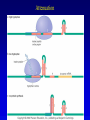
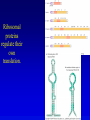
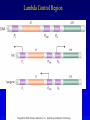
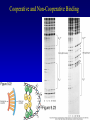
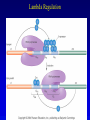
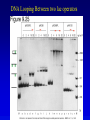
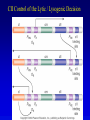
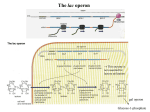
Chapter 16 Regulation in Prokaryotes 23 and 25 October, 2006 • • • • • • • • • • • • • Overview Transcriptional initiation is the most common point to regulate gene expression. Any of the events of initiation, including polymerase binding and open complex formation may be regulated either positively or negatively. Regulation is accomplished by sequence-specific DNA binding proteins. Binding may be promoter proximal or at a distance. DNA footprinting and mobility shift assays are used to investigate the binding of regulatory proteins. In the E. coli lac operon, there are both repressors and activators, each of which is allosterically regulated. Many regulatory systems control a large number of genes and operons, like the catabolite repression and heat shock regulons. NtrC is regulated by covalent modification, binds at a distance, and hydrolyzes ATP to pronmote open complex formation. MerR activates transcription by twisting the promoter. Riboswitches regulate transcription or translation without protein mediators. Phage lambda uses alternative regulatory systems to control lytic or lysogenic growth. Repressor and Cro compete to determine lytic or lysogenic growth, in response to the stability of the CII protein. Downstream regulation in lambda involves antitermination. Activators and repressors may regulate binding of polymerase. Some activators regulate open complex formation. Cooperative Binding and Transcriptional Regulation at a Distance The lac operon lac operon regulation Control Regions and lac Operator Half-sites RNA polymerase can form open complexes even in the presence of the LacI protein. RNA polymerase interacts with promoter and CAP Helix-turn-helix Interactions with DNA CAP bends DNA Activator Bypass Lac repressor binds as a tetramer Genetic experiments with partial diploids elucidated the ideas behind regulation of gene expression. Regulation by Alternative s-Factors Regulation of GlnA by s-54 and NtrC. NtrC Acts at a Distance MerR Regulation AraC Regulation Arabinose relaxes loops, and the loops reform in the absence of competitor. Regulation of the trp operon Tryptophan Interaction with Trp Repressor Attenuation Ribosomal proteins regulate their own translation. Riboswitches regulate gene expression without regulatory proteins. Phage lambda Lambda Genome Lambda Control Region Lambda Repressor and Binding Sites Cooperative Binding Cooperative and Non-Cooperative Binding Lambda Regulation Negative Autoregulation DNA Looping Between two lac operators CII Control of the Lytic / Lysogenic Decision N and Q Antiterminators int Regulation Title Title