* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Joining the Dots: Network Analysis of Gene Perturbation Screens
Oncogenomics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Public health genomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
RNA silencing wikipedia , lookup
Gene therapy wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Point mutation wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome evolution wikipedia , lookup
Gene nomenclature wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Microevolution wikipedia , lookup
RNA interference wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Designer baby wikipedia , lookup
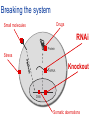
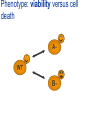
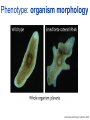
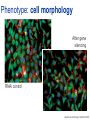
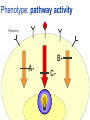
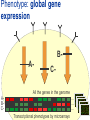
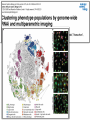
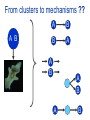
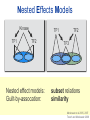
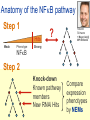
Joining the dots… Network analysis of gene perturbation data Florian Markowetz markowetzlab.org How to understand a complex system? M. mycoides JCVI-syn1.0 Richard Feynman: “What I cannot create, I do not understand.” Functional Genomics: “What I cannot break, I do not understand.” Breaking the system Drugs Small molecules RNAi Protein Stress mRNA Knockout DNA Somatic aberrations Today’s lecture • What information do we get out of gene perturbations? – Phenotypes and their ‘richness’ • How do we use this information to infer the internal architecture of a cell? – Guilt-by-association – Nested Effects Models Phenotype: viability versus cell death AWT B- Phenotype: organism morphology Boutros and Ahringer, Nat Rev 2008 Phenotype: cell morphology After gene silencing RNAi control Boutros and Ahringer, Nat Rev 2008 Phenotype: pathway activity Receptors BA- C- Phenotype: global gene expression B- A- C- All the genes in the genome … … … ABC- Transcriptional phenotypes by microarrays Phenotyping produces partslists Keith Haring, Untitled, 1986 Urs Wehrli, Tidying Up Art, 2003 A challenge for computation and statistics From phenotypes to clusters A B C A B From clusters to mechanisms ?? A B A B B A A B A B A B Nested Effects Models Kinase TF1 TF1 TF2 Nested effect models: Guilt-by-assocation: TF2 TF3 subset relations similarity Markowetz et al 2005, 2007 Tresch and Markowetz 2008 Nested Effects Models INPUT 1. Set of candidate pathway genes 2. High-dimensional phenotypic profile, e.g. microarray Gene perturbations OUTPUT Graph explaining the phenotypes Phenotypic profiles A B C D E F G H Inferred pathway AB EF CD GH Effects Anatomy of the NFB pathway Step 1 Hits Weak Phenotype ? Roland Schwarz + Meyer lab @ MPI IB Berlin Strong NFB Step 2 Knock-down Known pathway members New RNAi Hits Compare expression phenotypes by NEMs Nested Effect Models for NFB Roland Schwarz Take-home messages • Phenotyping screens probe a cell’s reaction to targeted perturbations • Guilt-by-assocation is a powerful predictor of gene/protein function … • … but Guilt-by-assocation has limited ability to infer mechanisms • Inferring subset relations by Nested Effects Models provides hierarchical view of cellular organisation PLoS Comput Biol 6(2) 2010 the team Joining the dots … Thank you ! Florian Markowetz markowetzlab.org