* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 11 Transcription and RNA Processing
Alternative splicing wikipedia , lookup
Molecular cloning wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Transposable element wikipedia , lookup
Gene regulatory network wikipedia , lookup
Transcription factor wikipedia , lookup
DNA supercoil wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic code wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
RNA interference wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Biosynthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Messenger RNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Polyadenylation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
RNA silencing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene expression wikipedia , lookup
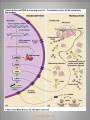
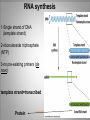
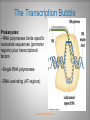
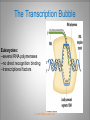
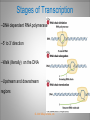
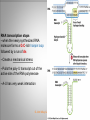
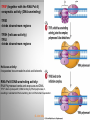
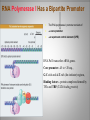
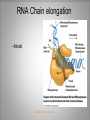
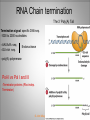
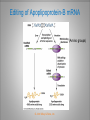
Chapter 11 Transcription and RNA Processing © John Wiley & Sons, Inc. Chapter Outline Transfer of Genetic Information: The Central Dogma The Process of Gene Expression Transcription in Prokaryotes Transcription and RNA Processing in Eukaryotes Interrupted Genes in Eukaryotes: Exons and Introns Removal of Intron Sequences by RNA Splicing © John Wiley & Sons, Inc. Transfer of Genetic Information: The Central Dogma Transfer of Genetic Information:The Central Dogma The central dogma of biology is that information stored in DNA is transferred to RNA molecules during transcription and to proteins during translation. DNA RNA Genotyping RNA proteins Phenotyping DNA/RNA virus © John Wiley & Sons, Inc. proteins The Central Dogma Transcription --one strand of DNA to one complementary strand of RNA --Uracil replaces Thymine (U-A bp) --primary transcripts messenger RNA (mRNA) -----chemical modification (splice=remove introns) (spliceosomes) © John Wiley & Sons, Inc. The Central Dogma Translation --RNA nucleotide into amino acids UUU--Phe ( F) phenylalanine(*) --genetics code (codons) --Ribosomes (ribonucleoproteins) (*) sense codon Non sense codon=stop codon © John Wiley & Sons, Inc. Transcription and Translation in Prokaryotes The primary transcript is equivalent to the mRNA molecule. The mRNA codons on the mRNA are translated into an amino acid sequence by the ribosomes. Retroviruses © John Wiley & Sons, Inc. Transcription and Translation in Eukaryotes The primary transcript (premRNA) is a precursor to the mRNA. The pre-mRNA is modified at both ends, and introns are removed to produce the mRNA. 3 After processing, the mRNA is exported to the cytoplasm for translation by ribosomes. © John Wiley & Sons, Inc. Types of RNA Molecules Messenger RNAs (mRNAs)—intermediates that carry genetic information from DNA to the ribosomes. Transfer RNAs (tRNAs)—adaptors between amino acids and the codons in mRNA. Ribosomal RNAs (rRNAs)—structural and catalytic components of ribosomes. © John Wiley & Sons, Inc. Types of RNA Molecules Small nuclear RNAs (snRNAs)—s tructural components of spliceosomes. Micro RNAs (miRNAs)—short single-stranded RNAs (20 to 22 bp) that block expression of complementary mRNAs. RNAi is similar to miRNA (RNA interference, double strand RNA, plant) siRNA (small interference) piRNA (Piwi-interacting RNA) is a small non-coding RNA molecules © John Wiley & Sons, Inc. The Central Dogma • The central dogma of molecular biology is that genetic information flows -----from DNA to DNA during chromosome replication, ----from DNA to RNA during transcription, ----from RNA to protein during translation. © John Wiley & Sons, Inc. Transcription involves the synthesis of a single strand RNA transcript complementary to one strand of DNA of a gene. • Translation is the conversion of information stored in the sequence of nucleotides in the RNA transcript into the sequence of amino acids in the polypeptide gene product, according to the specifications of the genetic code. © John Wiley & Sons, Inc. © John Wiley & Sons, Inc. The Process of Gene Expression The Process of Gene Expression For non-viral proteins… Information stored in the nucleotide sequences of genes is translated into the amino acid sequences of proteins through unstable intermediaries called messenger (m)RNAs. Synthesis of viral proteins… in infected bacteria involved an unstable RNA molecule synthesized from the viral DNA. © John Wiley & Sons, Inc. RNA synthesis 1-Single strand of DNA (template strand) 2-ribonuleoside triphosphate (NTP) 3-no pre-existing primers (de novo) template strand=transcribed Protein © John Wiley & Sons, Inc. Nucleophilic attacks --3’ hydroxyl group of the RNA strand --nucleotidyl phosphorus on the nucleoside triphosphate “nucleotide, nucleoside monophosphate” “RNA polymerase” “Transcriptional factors” © John Wiley & Sons, Inc. The Transcription Bubble Prokaryotes: --RNA polymerase binds specific nucleotide sequences (promoter regions) plus transcriptional factors --Single RNA polymerase --DNA unwinding (AT regions) © John Wiley & Sons, Inc. The Transcription Bubble Eukaryotes: --several RNA polymerases --no direct recognition binding --transcriptional factors © John Wiley & Sons, Inc. General Features of RNA Synthesis Similar to DNA Synthesis except – The precursors are ribonucleoside triphosphates. – Only one strand of DNA is used as a template. – RNA chains can be initiated de novo (no primer required). The RNA molecule will be complementary to the DNA template (antisense) strand and identical (except that uridine replaces thymidine) to the DNA non-template (sense) strand. RNA synthesis is catalyzed by RNA polymerases and proceeds in the 5’3’ direction. © John Wiley & Sons, Inc. • In eukaryotes, genes are present in the nucleus, whereas polypeptides are synthesized in the cytoplasm. • Messenger RNA molecules function as non-stable intermediaries that carry genetic information from DNA to the ribosomes, where proteins are synthesized. • RNA synthesis, catalyzed by RNA polymerases, is similar to DNA synthesis in many respects. • RNA synthesis occurs within a localized region of strand separation (Transcription Bubble), and only one strand of DNA functions as a template for RNA synthesis. © John Wiley & Sons, Inc. • RNA synthesis, catalyzed by RNA polymerases, is similar to DNA synthesis in many respects. Prokaryotic: OriC (245 bp) AT-rich region (replication bubble) Eukaryotic: ARS (Autonomously Replicating Sequences) AT-rich region 11 bp T O T cDNA 5’ 3’ 5’ 3’ http://www.youtube.com/watch?v=mFh9L-nu8Hk http://www.youtube.com/watch?v=OnuspQG0Jd0 Transcription in Prokaryotes Transcription ---The first step in gene expression ---Transfers the genetic information stored in DNA (genes) into messenger (m)RNA molecules that ---Carry the information to the sites of protein synthesis in the cytoplasm. © John Wiley & Sons, Inc. Stages of Transcription --DNA dependent RNA polymerase --5’ to 3’ direction --Walk (literally) on the DNA --Upstream and downstream regions © John Wiley & Sons, Inc. E. Coli RNA Polymerase Tetrameric core: 2 ’ Holoenzyme: 2 ’ (480,000 Daltons; bp~650 Daltons) Functions of the subunits: : assembly of the tetrameric core : ribonucleoside triphosphate binding site ’: DNA template binding region (sigma factor): initiation of transcription (*) (*) in vivo In vitro: RNA polymerase works…just fine on both DNA strands Initiation of RNA Chains Binding of RNA polymerase holoenzyme to a promoter region in DNA ( promoter region) Localized unwinding of the two strands of DNA by RNA polymerase to provide a single-stranded template Formation of phosphodiester bonds between the first few ribonucleotides in the nascent RNA chain © John Wiley & Sons, Inc. A Typical E. coli Promoter ..,-2,-1,+1,+2,.. © John Wiley & Sons, Inc. Numbering of a Transcription Unit The transcript initiation site is +1 (A/T). Bases preceding the initiation site are given minus (–) prefixes and are referred to as upstream sequences. Bases following the initiation site are given plus (+) prefixes and are referred to as downstream sequences. Consensus sequences: highly conserved Recognition sequences: Sigma factor ( © John Wiley & Sons, Inc. Elongation Sigma factor needs to be released ---Re- and Un-winding activities -- Walk (literally) on the DNA 5’ to 3’ --growing RNA chain RNA polymerase binds both DNA template and growing RNA chain © John Wiley & Sons, Inc. Termination Signals in E. coli Rho-dependent terminators—require a protein factor () Rho-independent terminators—do not require © John Wiley & Sons, Inc. Rho-independent terminators—do not require intrinsic termination) © John Wiley & Sons, Inc. RNA transcription stops --when the newly synthesized RNA molecule forms a G-C-rich hairpin loop followed by a run of As --Create a mechanical stress --Pulls the poly-U transcript out of the active site of the RNA polymerase --A-U has very weak interaction © John Wiley & Sons, Inc. Termination Signals in E. coli Rho-dependent terminators (non-intrinsic) — require a protein factor () and rut site Rut proteins bind specific RNA sequences (>>Cs and <<<Gs) Not hairpins or other secondary Structures Rho utilization (rut) © John Wiley & Sons, Inc. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIF F (Uncompressed) decompressor are needed to see this picture. Coupled Transcription and Translation in E. coli © John Wiley & Sons, Inc. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. stop (aka "nonsense") codon QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Rho factor binds to specific regions in naked RNA QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. • RNA synthesis occurs in three stages: (1) initiation, (2) elongation, and (3) termination. • RNA polymerases—the enzymes that catalyze transcription—are complex multimeric proteins. • The covalent extension of RNA chains occurs within locally unwound segments of DNA. © John Wiley & Sons, Inc. • Chain elongation stops when RNA polymerase encounters a transcription-termination signal. • Transcription, translocation, and degradation of mRNA molecules often occur simultaneously in prokaryotes. RNAase © John Wiley & Sons, Inc. Transcription and RNA Processing in Eukaryotes Five different enzymes catalyze transcription in eukaryotes, and the resulting RNA transcripts undergo three important modifications, including the excision of noncoding sequences called introns. The nucleotide sequenced of some RNA transcripts are modified post-transcriptionally by RNA editing. © John Wiley & Sons, Inc. Modifications to Eukaryotic pre-mRNAs A 7-Methyl guanosine cap is added to the 5’ end of the primary transcript by a 5’-5’ phosphate linkage. ( stability and protection) A poly(A) tail (a 20-200 nucleotide polyadenosine tract, As) is added to the 3’ end of the transcript. The 3’ end is generated by cleavage rather than by termination. (stability and protection) When present, intron sequences are spliced out of the transcript. (stability) © John Wiley & Sons, Inc. © John Wiley & Sons, Inc. Eukaryotes Have Five RNA Polymerases RNA polymerase II Nucleus miRNA Pre-mRNA~Heterogeneous nuclear RNA (hnRNA) © John Wiley & Sons, Inc. A Typical RNA Polymerase II Promoter (mRNA) Promoter: short sequence of conserved elements (seq. of DNA) located upstream from the transcript starting point. --~200 bp ( DNA linear) --~10 Kdp ( DNA bending) © John Wiley & Sons, Inc. Initiation by RNA Polymerase II Transcriptional factors (proteins) --Help/modulate/assist Basal transcriptional factors --bind close to the transcript starting point Other factors (enhancers and silencers) -TFIID --TATA-biding proteins (TBP) -TFIIA -TFIIB Where is the enzyme? © John Wiley & Sons, Inc. -TFIIF (together with the RNA Pol-II) --enzymatic activity (DNA-unwinding) -TFIIE --binds downstream regions -TFIIH (helicase activity) -TFIIJ --binds downstream regions Helicase activity: it separates two annealed nucleic acid strands RNA Pol-II DNA-unwinding activity: RNA Polymerase bends and wraps around DNA. TFIIF alters (nonspecific) DNA binding by RNA polymerase II, resulting in substantial DNA unwinding but not DNA strand separation. © John Wiley & Sons, Inc. -TFIIH (helicase activity and kinase activity) When RNA polymerase II binds to the complex, it initiates transcription. Phosphorylation of the CTD is required for elongation to begin. CTD: carboxy-terminal domain All eukaryotic RNA polymerases have ∼12 subunits and are aggregates of >500 kD. (nucleotide pair~0.660 kD) Some subunits are common to all three RNA polymerases. The largest subunit in RNA polymerase II has a CTD (carboxy-terminal domain) consisting of multiple repeats of a heptamer. -Typical RNA polymerase isolated from yeast (S. cerevisiase) ( and subunits) - subunits: CTD – carboxy-terminal domain, which consists in multiple repeats of 7 amino acids, unique and important of regulation (tyrosine (Try, Y), serine (Ser, S) and threonine (Thr, T) residues) -Some subunits are common to all three polymerases. Figure 24.2 RNA Polymerase I Has a Bipartite Promoter The RNA polymerase I promoter consists of: --a core promoter --an upstream control element (UPE) RNA Pol I transcribes rRNA genes. Core promoter: -45 to +20 seq., G-C-rich and A-T-rich (Inr-initiator) regions, Binding factors - protein complexes formed by TFIs and TBP-(TATA binding protein) RNA Polymerase III Uses Both Downstream and Upstream Promoters RNA polymerase III has (3) types of promoters. -RNA Pol III transcribes tRNA -Core promoters (boxes) -Transcriptional Factors(TF) III: general and specifics *proximal sequence element * Figure 24.7 RNA Chain elongation --Model © John Wiley & Sons, Inc. The 7-Methyl Guanosine (7-MG) Cap Histones:? FACT (facilitates chromatin transcriptional) Energy © John Wiley & Sons, Inc. RNA Chain termination The 3’ Poly(A) Tail Termination signal: specific DNA seq. -1000 to 2000 nucleotides --AAUAAA seq. --GU-rich seq. Endonuclease --poly(A) polymerase Pol-II vs Pol I and III -Terminator proteins (Rho-indep. Terminator) © John Wiley & Sons, Inc. RNA Editing Usually the genetic information is not altered in the mRNA intermediary. Sometimes RNA editing changes the information content of genes by – Inserting or deleting uridine monophosphate residues. – Changing the structures of individual bases © John Wiley & Sons, Inc. Editing of Apoplipoprotein-B mRNA (Amino groups) © John Wiley & Sons, Inc. • Three to five different RNA polymerases are present in eukaryotes, and each polymerase transcribes a distinct set of genes. • Eukaryotic gene transcripts usually undergo three major modifications: • • • the addition of 7-methyl guanosine caps to 5’ termini, The addition of poly(A) tails to 3’ ends, The information content of some eukaryotic transcripts is altered by RNA editing, which changes the nucleotide sequences of transcripts prior to their translation. © John Wiley & Sons, Inc. Interrupted Genes in Eukaryotes: Exons and Introns Most eukaryotic genes contain noncoding sequences called introns that interrupt the coding sequences, or exons. The introns are excised from the RNA transcripts prior to their transport to the cytoplasm. © John Wiley & Sons, Inc. Hybridization: annealing © John Wiley & Sons, Inc. R-Loop Evidence of an Intron in the Mouse -Globin Gene mRNA Missing in actions Pre-mRNA © John Wiley & Sons, Inc. Introns Introns (or intervening sequences) are noncoding sequences located between coding sequences. Introns are removed from the pre-mRNA and are not present in the mRNA. Introns are variable in size and may be very large ( 50 bp to 3000 bp). Exons (both coding and noncoding sequences) are composed of the sequences that remain in the mature mRNA after splicing. The biological significance of introns is still open to debate. © John Wiley & Sons, Inc. Removal of Intron Sequences by RNA Splicing The noncoding introns are excised from gene transcripts by several different mechanisms. Eukaryotes No prokaryotes (excepts a few a prokaryotes virus and others) © John Wiley & Sons, Inc. Excision of Intron Sequences Conserved seq. for mRNA Exon-GT…AG-exon intron 99% Ribonucleoproteins: Spliceosomes (1981) © John Wiley & Sons, Inc. Splicing Removal of introns must be very precise. Conserved sequences for removal of the introns of nuclear mRNA genes are minimal. – Dinucleotide sequences at the 5’ and 3’ ends of introns. Exon-GU…AG-exon – TACTAAC box (branch site with A) about 30 nucleotides upstream from the 3’ splice site. © John Wiley & Sons, Inc. Spliceosomes: snRNA plus ~40 proteins 1%: CG…AG AT…AC © John Wiley & Sons, Inc. Nuclear splicing involves transesterification GU…UACUAAC….AG “Branch site” Does splicing require energy? Does splicing ever occur in DNA? Figure 26.7 tRNA A splicing endonuclease makes two cuts at the end of the intron. A splicing ligase joins the two ends of the tRNA to produce the mature tRNA. Specificity resides in the three-dimensional (secundary) structure of the tRNA precursor, not in the nucleotide sequence. © John Wiley & Sons, Inc. rRNA (Autocatalytic Splicing) G-3’-OH: absolute requirement “Co-factor” © John Wiley & Sons, Inc. • Noncoding intron sequences are excised from RNA transcripts in the nucleus prior to the transport to the cytoplasm. • Introns in tRNA precursors are removed by the concerted action of a splicing endonuclease and ligase, whereas introns in some rRNA precursors are spliced out autocatalytically—with no catalytic protein involved. © John Wiley & Sons, Inc. • The introns in nuclear pre-mRNAs are excised on complex ribonucleoprotein structures called spliceosomes. • The intron excision process must be precise, with accuracy to the nucleotide level, to ensure that codons in exons distal to introns are read correctly during translation. © John Wiley & Sons, Inc.