* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Idling behind the Yellow Line: Cybercensorship and the Liability of
Survey
Document related concepts
Two-hybrid screening wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Genetic code wikipedia , lookup
Protein–protein interaction wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Signal transduction wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Biochemistry wikipedia , lookup
Proteolysis wikipedia , lookup
Protein structure prediction wikipedia , lookup
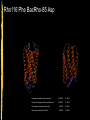
SNARE (protein) wikipedia , lookup
Transcript
Property Based Conservation Percolation in Transmembrane Proteins JKS SEMINAR Oznur Tastan November 21, 2005 1 PERCOLATION 2 Percolation Threshold, pc Percolation theory deals with the connectivity of components a system. p< pc there is no infinite cluster p> pc there is one infinite cluster White balls => nonconducting spheres Black ball =>conducting spheres . Only above a certain concentration pc of the conducting balls the system conducts electrical current. 3 Membrane Folding Klein-Seetharaman,J., 2005 4 So far… Three very initial points have been explored: 1. Membrane assignment of residues. 2. Build a data set of up-to-date membrane proteins of structure known. 3. The calculation of hydrophobic percentage of membrane proteins 4. Initial program of percolation 5 PDBTM PDB-TM is a new database for tm proteins with known structures (Tusnady et. al. 2004). * weekly updated http://pdbtm.enzim.hu/ Rhodopsin 1u19, A Membrane approximation and oriented structures with respect to the menbrane 6 Membrane proteins compiled There are 179 chains in the non-redundant alpha list of PDB-TM Followings are excluded: structures with less than three transmembrane helices. structures unusual very open theoretical models 54 membrane protein chains 7 Hydrophobic percentage The three differing amino acids for each set: All 6 hydrophobicity sets share the same six amino acids: Ile, Leu, Val, Phe, Trp, Met. Set 1 Set 1 [Cys,His,Tyr] Set 2 [Cys,Pro,Tyr] Set 3 [Cys,Ala,Tyr] Set 4 [Pro,Ala,Tyr] Set 5 [Cys,Ala,Gly] Set 6 [Pro,Ala,Gly] Set 2 Set 3 Set 4 Set 5 Set 6 Membrane <fprot>54 0.47 0.50 0.55 0.58 0.60 0.64 Soluble <fprot>103 0.35 0.37 0.42 0.44 0.47 0.49 As expected membrane proteins contains more hydrophobic residues on the average. 8 Finding clusters, a breadth first search Start from an occupied cell. Add the first neighbors of the origin to the cluster, Add the first neighbors of children Grow cluster until there is no neighbors Continue until there is no occupied cell Hydrophobic Clusters residues in Rhodopsin Connectivity cutoff(Å) Number of clusters Size of largest cluster End-to-end distance of the largest cluster(Å) 7 6 161 61.55 6 16 104 55.76 5 65 12 15.12 9 Largest cluster of hydrophobic residues at 7 6 Å cutoff The largest cluster of hydrophobic residues at 6 Å cutoff. 10 Property Based Conservation 11 Rhodopsin and MGluR6 PBC comparison 12 23 Properties selected 4 A contact number/Nishikawa-Ooi 8 A contact number/Nishikawa-Ooi Average accessible surface area/Janin Flexibility param for no rigid neighbors/Karplus-Schulz Flexibility param for one rigid neighbor/Karplus-Schulz Flexibility param for two rigid neighbors/Karplus Schulz7Hphob/Miyazawa/Roseman8 Hydropathy index/Kyte-Doolittle Long range non-bonded energy per atom/Oobatake-Ooi Normalized flexibility/B-values, average/Vihinen Normalized frequency of alpha-helix/Chou-Fasman Normalized frequency of beta-sheet/Chou-Fasman Normalized frequency of coil/Nagano Normalized frequency of turn/Crawford Normalized van der Waals volume/Fauchere Number of hydrogen bond donors/Fauchere Percentage of buried residues/Janin Polarity/Grantham Short and medium range non-bonded energy per residue/Oobatake-Ooi Side chain torsion angle phi/AAAR Levitt Volume/Grantham White Wimley Octanol Interface Scale Helix-PackingScale 13 Method The permutation test is not employed. Instead the results are normalized with background probabilities, which are calculated class specific, by taking the frequencies of amino acid in each of the alignments Positions that exhibit similar properties are identified. normalized conservation ratio, r normalized conservation difference, d consider similar behavior if|r-1| <0.05 or |d|<0.01 37 positions to be examined in the folding core 14 Rho116 Phe Mglur6-650 Leu 15 Rho116 Phe MGluR6-650 Leu 'Average accessible surface area/Janin' 0.075881 'Percentage of buried residues/Janin' 0.18762 'Hphob/Miyazawa/Roseman' 0.2938 'Flexibility param for two rigid neighbors/Karplus-Schulz' 0.33739 '8 A contact number/Nishikawa-Ooi' 0.39612 0.09896 0.25537 0.32786 0.42779 0.38902 16 Bacteriodopsin and Rhodopsin Green is bacteriorhodopsin Blue is rhodopsin 17 Rho116 Phe BacRho-85 Asp 18 Rho116 Phe BacRho-85 Asp 'Average accessible surface area/Janin' 0.075881 0.15427 'Number of hydrogen bond donors/Fauchere' 0.084191 0.14124 'Percentage of buried residues/Janin' 0.18762 0.16367 'Hydropathy index/Kyte-Doolittle' 0.23445 0.18368 19 T4 lyzosome mutations To be used in the PBC analysis. T4 lyzosome mutations are started to be compiled The full mutation list is obtained from ProTherm. There are 1068 entries. We collected all the available online papers that is related to the mutations. Summarizing the effect and the nature of the mutations 20 To be continued.. Thanks to Naveena, Judith, Jaime and Hagai 21