* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Kinetic proofreading - Weizmann Institute of Science
Survey
Document related concepts
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Genetic code wikipedia , lookup
Bottromycin wikipedia , lookup
Epitranscriptome wikipedia , lookup
Molecular cloning wikipedia , lookup
Molecular evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Biochemistry wikipedia , lookup
Expanded genetic code wikipedia , lookup
Point mutation wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transcript
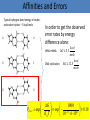
Kinetic proofreading tRNA – Ribosome analogy J.J. Hopfield 1974 Outline • High precision bio-synthetic processes • The matching problem and its solution by kinetic proofreading • Examples and more recent results tRNA-mRNA matching (protein synthesis) Remember: coding redundancy DNA replication Perror 10 9 Less than 1 error per strand (In human chromosome #1 there are ~200,000,000 base pairs ) Affinities and Errors Typical hydrogen bond energy of codonanticodon triplets ~ 5 kcal/mole A U In order to get the observed error rates by energy difference alone: tRNA-mRNA: G C kcal G 5.5 mole kcal DNA replication: G 12.5 mole kcal G AU GGU G ~ 1 mole K BT 1021 cal G U Perror G 1000 exp 21 exp 0.18 23 10 6 10 K BT Michaelis – Menten Kinetics ES Enzyme ES k 1 Substrates k1 Enzyme substrates complex PS E k2 Product d ES k1 E S k 1 k 2 ES dt Hopfield’s problem The desired enzymatic process The undesired enzymatic process k 'c C c Cc PC w kc k 'D D c Dc PD w kD Assumptions: C D c Steady state error rate is embodied in the reaction rates k 'C k 'D c c w - much smaller than the other rates G PD w kC kC e RT f 0 PC w k D k D Hopfield’s Solution k 'c C c Cc Cc PC m' * w kc lC C c m' k C k D w is negligible With these kinetics: Dc Cc f0 lC f0 And with: lD PD 2 f0 PC Another option: one step and time dependent reaction rates. Kinetic proofreading • • • • Multistep process. Discard step. Directionality by energy expenditure. Dominance of direct production. k 'c C c Cc kc Cc Pc * m, m' lC l'C C c w c Proofreading - Protein Synthesis GTP (Hopfield 1974) GDP+P Experimental result – Protein synthesis • • • • Blanchard et al. 2004 Fluorescently labeled tRNA molecules. Antibiotic inhibitors of tRNA selection. Nonhydrolizable GTP analogues. Enzymatically and chemically altered ribosome complexes GTPase activity stimulation Codon recognition (different rates, k3, for cognate state and non-cognate) GTP hydrolysis Phosphate releaseProofreading Experimental result – tRNA & amino acid binding Measuring concentrations in time of correct (isoleucine) and incorrect (valine) charged tRNAs Energy expenditure Correct / incorrect? DNA replication Additional step forward function of the enzyme (DNA polymerase) Schaaper 1993 Conclusions and Key Points Specificity through energetic differences isn’t enough. To achieve enzymatic proofreading: • Directionality through energy consumption • Discard steps. • Multi-steps. Living cells need to regulate substance concentration and control reaction rates to achieve the conditions for the nest proofreading chain.