* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Our Genes, Our Drugs and our Future
Survey
Document related concepts
Polysubstance dependence wikipedia , lookup
Orphan drug wikipedia , lookup
Compounding wikipedia , lookup
Psychopharmacology wikipedia , lookup
Theralizumab wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Drug design wikipedia , lookup
Pharmacognosy wikipedia , lookup
Pharmaceutical industry wikipedia , lookup
Neuropharmacology wikipedia , lookup
Prescription costs wikipedia , lookup
Drug discovery wikipedia , lookup
Drug interaction wikipedia , lookup
Transcript
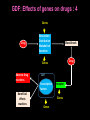
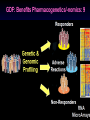
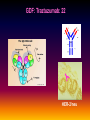
Pieter van der Bijl, Emeritus Professor and Former Head of Pharmacology, Faculty of Health Sciences, Stellenbosch University, Cape Town, South Africa GDF: Introduction: 1 The focus of traditional medical practice is on clinical signs and symptoms in accordance with medical history Not always the most effective approach given the different genetic profile of each individual Pharmacogenetic studies over many decades have documented that genetic variability can affect PK and PD Polymorphisms of drug metabolising enzymes, transporters and receptors contribute to variable drug responses (in addition to environmental , physiological & compliance factors) GDF: Introduction: 2 Most drugs act on: Enzymes Transporters Membrane ion channels Receptors and Are biotransformed by: Drug metabolising enzymes Drug GDF: Introduction: 3 Foregoing molecular systems are all proteins coded for by certain genes Not surprising that genetic factors are major determinants of variability of drug effects and many pts do not respond adequately to their medication ( confusion in the‘therapeutic jungle’) Some blockbuster drugs have only limited efficacy in 70% of pts Many reasons for this, but pharmacogenetics plays an important role Most information hitherto from genetic diversity obtained wrt drug metabolising enzymes GDF: Effects of genes on drugs : 4 Genes Drug Absorption Distribution Metabolism Excretion Blood levels Drug Genes Adverse drug reactions Cell Transcription factors Beneficial effects reactions Receptor Genes Genes GDF: Variable efficacy of drugs: 5 Drug Class Insufficient response (%) SSRIs 10-25 ACE-inhibitors 10-30 -blockers 15-25 Antidepressants 20-50 Statins 30-70 2-agonists 40-70 GDF: Pharmacogenetics/-nomics: 6 Recently pharmacogenetics has evolved into ‘pharmacogenomics’ The latter involves a shift from a focus on individual candidate genes to genome-wide association studies Pharmacogenomics is a precursor of personalised medicine This constitutes a shift from ‘one-drug-fitsall’ to ‘the right drug for the right patient at the right dose and time’ But, each pt will not be treated differently from every other pt (economically untenable) GDF: Pharmacogenetics/-nomics: 7 Rather, pts will be divided into groups by genetic and other markers that predict disease progression and treatment outcome For drug treatment one needs to avoid lack of response or toxicity If ADRs can be from 5% to 2% by excluding 10% of the targeted population, the drug gains a better risk/benefit ratio 1st choice Rx and a market share There is a growing trend to link new drugs with diagnostic biomarkers (often genetic) (FDA, EMA) improved Rx outcome (personalised medicine!) GDF: Benefits Pharmacogenetics/-nomics: 8 Improvement of drug choices In USA 100 000 pts die annually due to ADRs and 2 000 000 are hospitalised Pharmacogenomics will predict who will have a + or reaction Safer dosing options Testing of genomic variation will correct dosing Improved drug development industry to determine in which populations new drugs will be effective Decreased health care costs deaths and hospitalisation due to ADRs purchase of drugs ineffective in certain pts Speed up clinical trials for new drugs GDF: Benefits Pharmacogenetics/-nomics: 9 GDF: Altered drug responses: 10 P-genetic biomarker Drug Disease FDA class. EMA labels Aim of genotyping Elimination of ADRs G6PD deficiency Primaquine Malaria + NAT variants INH Tuberculosis + * Elimination of ADRs CCR5 expression Maraviroc HIV +++ ** Efficiency C-KIT expression Imatinib GI stromal tumour + ** Efficiency CYP2C9 & VKORC1 variants Warfarin Thromboembolism ++ Elimination of ADRs CYP2C19 variants Voriconazole Fungal infection + Elimination of ADRs EGFR expression and KRAS mutation Cetucimab Colorectal CA +++ ** Efficiency HER/neu overexpression Traztuzumab Breast CA +++ ** Efficiency Ph1 chromosome Imatinib ALL +++ ** Efficiency TPMT variants Mercaptopurine ALL ++ * Efficiency UGT1A1 variants Irinotecan Colorectal CA ++ +++ required; ++ recommended; + for information only Elimination of ADRs ** included into indication or contraindication label * included in the other label information GDF: G6PD deficiency: 11 G6PD (glucose-6-phosphate dehydrogenase) an enzyme in hexose monophosphate shunt (a main source of NADPH generation) NADPH needed to reduce disulphide bonds of glutathione (GS-SGGSH) and other proteins Many drugs and their metabolites can use up GSH and lead to GSH levels in G6PD deficient pts GSH deficiency in RBC results in: Membrane fragilityhaemolysishaemolytic anaemia GDF: G6PD deficiency: 12 GDF: N-acetylation: 13 In late 1940’s discovered that there was a high incidence of peripheral neuropathy in pts Tx with isoniazid (INH) for tuberculosis INH is cleared from the blood after acetylation in the liver by N-acetyltransferase (NAT2) Hereafter the N-acetyl INH and some minor metabolites are excreted in the urine Hepatic insufficiency may t½ GDF: N-acetylation: 14 Acetyl CoA acts as a donor of the acetylgroup on INH N CO-NH-NH2 + CoA-COCH3 Isoniazid (INH) N N-Acetyltransferase (NAT2) Acetyl-Coenzyme A CO-NH-NH-COCH3 + CoA Acetyl-Isoniazid (INH) Coenzyme A GDF: Rapid and slow acetylators: 15 Individuals who are rapid acetylators: Have failure rate with INH in Tx of TB Require doses of hydralazine to control HT Individuals who are slow acetylators have risk of: Drug-induced SLE with hydralazine Haematological ADRs after INH Idiosyncratic ADRs to sulphonamides Bladder CA after exposure to carcinogenic arylamines Breast CA in postmenopausal females (4x) GDF: N-acetylation: 16 Work done in the Department of Pharmacology, SU has shown that there exist three well-defined groups of acetylators of INH [N-acetyltransferase (NAT2)] in the Western Cape Coloured population (mixed-race) The proportion of patients in these groups (fast, intermediate and slow) depends on racial characteristics I 50% Wide variation in other ethnic populations found S 30% (Eskimo’s, Asians, Africans, European & Egyptian) F 20% GDF: Warfarin: 17 The most commonly prescribed anticoagulant (vit K antagonist) (role of dabigatran & rivaroxaban) Has a narrow therapeutic index that varies widely between individuals (monitoring) Pts may be: Resistant and need dose to prevent CVAs (strokes) Sensitive and need dose to prevent CNS bleeding Metabolised by CYP450 (CYP2C9*2 and *3) Vit K is recycled by vit K epoxide reductase (VKORC1) GDF: Warfarin: 18 GDF: Genomics/Proteomics: 19 Implications of postgenomic medicine wrt drug development Apart from improved drug choices and development, safer dosing and decreased health costs genetically modified organisms can be used for drug production, eg insulin, monoclonal antibodies etc GDF: Genomics/Proteomics: 20 GDF: Genomics/Proteomics: 21 First regulatory review of targeted therapeutic agent with diagnostic test Approval of trastuzumab and HercepTest for pts with HER-2/neu overexpressing in breast CA (FDA,1999) Trastuzumab (Herceptin) is a humanised IgG1 against ectodomain of the HER-2 /neu receptor GDF: Traztuzumab: 22 HER-2/neu GDF: Genomics/Proteomics: 23 Another example, diagnostic kit for Bcr-Abl translocation in CML and selection for Rx with small molecule drug, imatinib (Gleevec) Acts by inhibiting tyrosine kinase and activation of target proteins in cellular proliferation GDF: Imatinib: 24 N H N N H3C O H N CH3 N N N Tyrosine kinase GDF: Genomics/Proteomics: 25 Also variety of diagnostic tests for management of major nonmalignant diseases are becoming available Germline-based SNP detection or biomarkers on serum or synovial fluid for progression of RA and selection of Rx Measurement of C-reactive protein markers in novel ways for CV disease Also genotyping and molecular diagnostics for diabetes mellitus Viral load testing and drug resistance (HIV) measuring is becoming standard GDF: Genomics/Proteomics: 26 Conclusions Trial and error medicine to precise biomarker-assisted diagnosis and more effective molecularly-guided Rx For drug companies efficiency, productivity and product lines Over next 5 years will see large impact of targeted drug approach guided by diagnostic tests in Rx of CA Exciting future wrt new specific drug development increased quality of life GDF: Personalised prescribing- the future: 27 1. mRNA extracted from sample 2. cDNA copies made and dye (green/red)labelled (eg CA/N) 3. Microarray, 1000’s wells (many identical copies of same gene) 4. cDNA pipetted into each well and hybridizes with complementary strands (wash) 5. Microarray scanner 6. Expression pattern obtained •A drop of blood or smear from buccal pouch •Microchip (gene chip) checks for 31 variations (polymorphisms) in two genes (CYP2D6 & CYP2C19) •Phenotype (eg ultrarapid metabolisers) identified ?