* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download CYP 2D6 Polymorphism
Survey
Document related concepts
Restriction enzyme wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Metabolomics wikipedia , lookup
Biochemical cascade wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Drug design wikipedia , lookup
Drug discovery wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Endocannabinoid system wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Lipid signaling wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Signal transduction wikipedia , lookup
Pharmacometabolomics wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Transcript
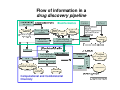
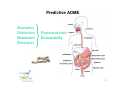
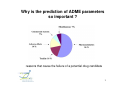
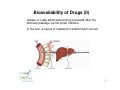
Flow of information in a drug discovery pipeline Bioinformatics Computational and Combinatorial Chemisty 1 Predictive ADME Absorption Distribution Metabolism Elimination Pharmacokinetic Bioavailability 2 Why is the prediction of ADME parameters so important ? reasons that cause the failure of a potential drug candidate 3 Bioavailablity of Drugs (I) 4 Bioavailability of Drugs (II) Uptake of orally administered drug proceeds after the stomach passage via the small intestine. In the liver, a series of metabolic transformation occurs. 5 Cytochrome P450 The super-family of cytochrome P450 enzymes has a crucial role in the metabolism of drugs. Almost every drug is processed by some of these enzymes. This causes a reduced bioavailability. Cytochrome P450 enzymes show extensive structural polymorphism (differences in the coding region). 6 Cytochrome P450 metabolisms (I) During first liver passage: First pass effect extensive chemical transformation of lipophilic or heavy (MW >500) compounds. They become more hydrophilic (increased water solubility) and are therefore easier to excreat. H CH3 O COOH phase I N COOH phase II Predominantly cytochrome P450 (CYP) enzymes are responsible for the reactions belonging to phase I. Usually, the reaction is a monooxygenation. 7 Cytochrome P450 Metabolismus (II) The substrates are monooxygenated in a catalytic cycle. Drug-R + O2 CYP NADPH Drug-OR + H2O NADP The iron is part of a HEM moiety 8 Cytochrome P450 Metabolismus (III) The cytochromes involved in the metabolism are mainly monooxygenases that evolved from the steroid and fatty acid biosynthesis. So far, 17 families of CYPs with about 50 isoforms have been characterized in the human genome. classification: CYP 3 A 4 *15 A-B family isoenzyme allel >40% sequencesub-family homology >55% sequencehomology 9 Cytochrome P450 gene families Human 14+ Molluscs 1 CYP450 Plants 22 Insects 3 Fungi 11 Bacteria 18 Yeasts 2 Nematodes 3 10 Human cytochrome P450 family Of the super-family of all cytochromes, the following families were confirmed in humans: CYP 1-5, 7, 8, 11, 17, 19, 21, 24, 26, 27, 39, 46, 51 Function: CYP 1, 2A, 2B, 2C, 2D, 2E, 3 metabolismus of xenobiotica CYP 2G1, 7, 8B1, 11, 17, 19, 21, 27A1, 46, 51 steroid metabolism CYP 2J2, 4, 5, 8A1 fatty acid metabolism CYP 24 (vitamine D), 26 (retinoic acid), 27B1 (vitamine D), ... 11 Cytochrome P450 enzymes (I) Flavin Monooxygenase Isoenzyme Alkohol Dehydrogenase Aldehyd Oxidase Monoamin Dehydrogenase (MAO) Drug-R + O2 CYP NADPH The redox activity is mediated by an iron porphyrin in the active center Drug-OR + H2O NADP 12 Cytochrome P450 enzymes (II) Despite the low sequence identity between CYPs from different organisms, the tertiary structure is highy conserved. Superposition of hCYP 2C9 (1OG5.pdb) and CYP 450 BM3 (2BMH.pdb) Bacillus megaterium In contrast to bacterial CYPs, the microsomal mammalian CYPs possess an additional transmembrane helix that serves as an anchor in the membrane 13 Cytochrome P450 enzymes (III) The structures of several mammalian CYPs have now been determined in atomistic detail and are available from the Brookhaven Database: http://www.pdb.mdc-berlin.de/pdb/ 1DT6.pdb CYP 2C5 rabbit Sep 2000 1OG5.pdb CYP 2C9 human Jul 2003 1PO5.pdb CYP 2B4 rabbit Oct 2003 1PQ2.pdb CYP 2C8 human Jan 2004 They are suitable templates for deriving homology models of further CYPs 14 Cytochrome P450 enzymes (IV) The majority of CYPs is found in the liver, but certain CYPs are also present in the wall cells of the inestine The mammalian CYPs are bound to the endoplasmic reticulum, and are therefore membrane bound. CYP 2D6 2% CYP 2A6 4% CYP distribution other 7% CYP 3 31% CYP 1A2 13% CYP 1A6 8% CYP 2C6 6% CYP 2E1 13% CYP 2C11 16% CYP 3 CYP 2C11 CYP 2E1 CYP 2C6 CYP 1A6 CYP 1A2 CYP 2A6 CYP 2D6 other 15 Cytochrome P450 enzymes (V) Especially CYP 3A4, CYP 2D6, and CYP 2C9 are involved in the metabolism of xenobiotics and drugs. hepatic only Metabolic Contribution CYP 2C9 10% CYP 1A2 other 2% 3% CYP 3A4 CYP 2D6 CYP 2C9 CYP 1A2 other CYP 3A4 55% CYP 2D6 30% also small intestine 16 Substrate specificity of CYPs (I) spezific substrates of particular human CYPs CYP 1A2 verapamil, imipramine, amitryptiline, caffeine (arylamine N-oxidation) CYP 2A6 nicotine CYP 2B6 cyclophosphamid CYP 2C9 diclofenac, naproxen, piroxicam, warfarin CYP 2C19 diazepam, omeprazole, propanolol CYP 2D6 amitryptiline, captopril, codeine, mianserin, chlorpromazine CYP 2E1 dapsone, ethanol, halothane, paracetamol CYP 3A4 alprazolam, cisapride, terfenadine, ... see also http://medicine.iupui.edu/flockhart/ 17 Substrate specificity of CYPs (II) Decision tree for human P450 substrates CYP 1A2, CYP 2A-E, CYP 3A4 CYP 2E1 CYP 2C9 low Volume high medium acidic basic pK a CYP 3A4 CYP 2D6 neutral CYP 1A2, CYP 2A, 2B CYP 2B6 low planarity high CYP 1A2 medium CYP 2A6 Lit: D.F.V. Lewis Biochem. Pharmacol. 60 (2000) 293 18 Cytochrome P450 polymorphisms „Every human differs (more or less) “ The phenotype can be distinguished by the actual activity or the amount of the expressed CYP enzyme. The genotype, however, is determined by the individual DNA sequence. Human: two sets of chromosomes That mean: The same genotype enables different phenotypes Depending on the metabolic activity, three major cathegories of metabolizers are separated: extensive metabolizer (normal), poor metabolizer, and ultra-rapid metabolizer (increased metabolism of xenobiotics) Lit: K. Nagata et al. Drug Metabol. Pharmacokin 3 (2002) 167 19 CYP 2D6 Polymorphism (I) The polymorphismus of CYP 2D6 (debrisoquine 4hydroxylase) has been studied in great detail, as metabolic differences have first been described for debrisoquine and sparteine (antipsychotics) localized on chromosome 22 Of the 75 allels, 26 exprime CYP2D6 proteines see http://www.imm.ki.se/CYPalleles/cyp2d6.htm 20 CYP 2D6 Polymorphism (II) Lit: J. van der Weide et al. Ann. Clin. Biochem 36 (1999) 722 21 CYP 2D6 Polymorphism (III) MGLEALVPLAVIVAIFLLLVDLMHRRQRWAARYPPGPLPLPGLGNLLHVDFQNTPYCFDQ poor debrisoquine metabolism S R impaired mechanism of sparteine LRRRFGDVFSLQLAWTPVVVLNGLAAVREALVTHGEDTADRPPVPITQILGFGPRSQGVF poor debrisoquine metabolism I LARYGPAWREQRRFSVSTLRNLGLGKKSLEQWVTEEAACLCAAFANHSGRPFRPNGLLDK poor debrisoquine metabolism R AVSNVIASLTCGRRFEYDDPRFLRLLDLAQEGLKEESGFLREVLNAVPVLLHIPALAGKV LRFQKAFLTQLDELLTEHRMTWDPAQPPRDLTEAFLAEMEKAKGNPESSFNDENLRIVVA missing in CYP2D6*9 allele DLFSAGMVTTSTTLAWGLLLMILHPDVQRRVQQEIDDVIGQVRRPEMGDQAHMPYTTAVI P loss of activity in CYP2D6*7 HEVQRFGDIVPLGMTHMTSRDIEVQGFRIPKGTTLITNLSSVLKDEAVWEKPFRFHPEHF LDAQGHFVKPEAFLPFSAGRRACLGEPLARMELFLFFTSLLQHFSFSVPTGQPRPSHHGV FAFLVSPSPYELCAVPR T impaired metabolism of sparteine in alleles 2, 10, 12, 14 and 17 of CYP2D6 see http://www.expasy.org/cgi-bin/niceprot.pl?P10635 22 CYP 2D6 Polymorphism (III) variability of debrisoquine-4-hydroxylation HO N NH2 NH H CYP2D6 NH2 N NH = number of individuals (european population) homocygote extensive metabolizers homocygote poor metabolizers = metabolic rate heterocygote extensive metabolizers Lit: T. Winkler Deutsche Apothekerzeitung 140 (2000) 38 23 Polymorphisms of further CYPs CYP 1A2 individual: fast, medium, and slow turnover of caffeine CYP 2B6 missing in 3-4 % of the caucasian population CYP 2C9 deficit in 1-3 % of the caucasian population CYP 2C19 individuals with inactive enzyme (3-6 % of the caucasian and 15-20 % of the asian population) CYP 2D6 poor metabolizers in 5-8 % of the european, 10 % of the caucasian, and <1% of the japanese population. Over expression (gene duplication) among parts of the african and oriental population. CYP 3A4 only few mutations 24 Genotyping for P450 alleles Affymetrix (US) has developped microarrays (gene chips) using immobilized synthetic copies of P450 nucleotides, that allow the identification of all clinically relevant allelic variants. 25 Induction and regulation of CYP3A (I) A series of xenobiotics have been identified, that lead to increased expression of enzymes of the CYP3A family. Indinavir efavirenz cyclosporin carbamazepine atorvastatin tamoxifen antiviral antiviral immuno-suppressant antispychotic HMG CoA Reductase Inhibitor anti-hormone These bind to the pregnane X receptor (PXR) which is the transcription factor for the regulation of the CYP3A gene expression. Lit: T.M. Wilson et al. Nature Rev. Drug Disc. 1 (2002) 259 26 Induction and regulation of CYP3A (II) The PXR receptor functions together with the retinoid X receptor (RXR) as a heterodimer. CYP3A induction leads to an increased metabolism of the administered substance due to upregulated enzymes. This can cause adverse reactions, like inflammation of the liver (hepatitis). 27 RXR and other nuclear receptors (I) As a specific, endogen activator of RXR, 5β-pregnane3,20-dione has been identified. In contrast, PXR is much less specific and is activated by glucocorticoids as well as by anti-glucocorticoids. Conversely, the unspecific constitutive androgen receptor (CAR) is found in the cytoplasm and dimerizes with PXR in the nucleus. Analog to PXR, the CYP2B gene is regulated. Likewise high sequence homology has been found for the vitamine D receptor (VDR) that regulates CYP27, and for the arylhydrocarbon receptor (AHR) (dioxin receptor). 28 RXR and other nuclear receptors (II) These nuclear receptors all belong to a family of transcription factors. Each one possess a double zinkfinger DNA-binding domain (DBD), and a larger ligand binding domain (LBD) which is carboxy terminal. They have been called orphan nuclear receptors as their ligands have been found later. 29 Nuclear Receptors as Drug Targets Contribution to the human genome and number of marketed drugs 30 Induction and regulatiion of CYP3A (III) O HO O O hyperforin, a natural ingredient of St. John‘s wort (Johanniskraut, Hypericum performatum) exhibits the highest measured affinity to PXR (Kd = 27 nM) so far. Application: remedy against cholestasis, mild antidepressant 31 Induction and regulation of CYP3A (IV) X-ray structure of PXR with bound hyperforin (1M13.pdb) Lit: R.E. Watkins et al. Biochemistry 42 (2003) 1430 32 Induction of further CYPs CYP 1A2 omeprazole, insulin, aromatic hydrocarbons (cigarette smoking, charbroiled meat) causes increased caffeine level in the plasma, if you quit smoking. CYP 2C9 rifampicin, secobarbital CYP 2C19 carbamazepine, prednisone CYP 2D6 dexamethason CYP 2E1 ethanol, isoniazid CYP 3A4 glucocorticoide, phenobarbitone, rifampicin, nevirapine, sulfadimindine, nevirapine, sulfinpyrazone, troglitazone 33 Typical inhibitors of various CYPs CYP 1A2 cimetidine, ciprofloxacine, enoxacine... grapefruit juice (naringin, 6‘,7‘-dihydroxybergamottin) CYP 2C9 chloramphenicol, amiodarone, omeprazole,... CYP 2C19 fluoxetine, fluvastatin, sertraline,... CYP 2D6 fluoxetine, paroxetine, quinidine, haloperidol, ritonavir,... CYP 2E1 disulfiram, cimetidine,... CYP 3A4 cannabinoids, erythromycin, ritonavir, ketokonazole, grapefruit juice see also http://medicine.iupui.edu/flockhart/ 34