* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Operons: The Basic Concept
Biochemistry wikipedia , lookup
Protein adsorption wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Genetic code wikipedia , lookup
Protein moonlighting wikipedia , lookup
RNA interference wikipedia , lookup
Gene expression profiling wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Expression vector wikipedia , lookup
Polyadenylation wikipedia , lookup
Molecular evolution wikipedia , lookup
Proteolysis wikipedia , lookup
Point mutation wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene regulatory network wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Messenger RNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene expression wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transcriptional regulation wikipedia , lookup
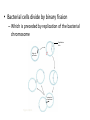
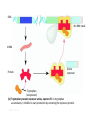
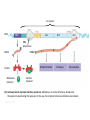
Operons: The Basic Concept • In bacteria, genes are often clustered into operons, composed of – An operator, an “on-off” switch – A promoter – Genes for metabolic enzymes • Bacterial cells divide by binary fission – Which is preceded by replication of the bacterial chromosome Replication fork Origin of replication Termination of replication Figure 18.14 Mutation and Genetic Recombination as Sources of Genetic Variation • Since bacteria can reproduce rapidly – New mutations can quickly increase a population’s genetic diversity • An operon – Is usually turned “on” – Can be switched off by a protein called a repressor • The trp operon: regulated synthesis of repressible enzymes trp operon Promoter DNA Promoter Genes of operon trpD trpC trpE trpR trpB trpA Operator Regulatory gene mRNA 5 3 RNA polymerase Start codon Stop codon mRNA 5 E Protein Inactive repressor D C B Polypeptides that make up enzymes for tryptophan synthesis (a) Tryptophan absent, repressor inactive, operon on. RNA polymerase attaches to the DNA at the promoter and transcribes the operon’s genes. Figure 18.21a A DNA No RNA made mRNA Protein Active repressor Tryptophan (corepressor) (b) Tryptophan present, repressor active, operon off. As tryptophan accumulates, it inhibits its own production by activating the repressor protein. Figure 18.21b Repressible and Inducible Operons: Two Types of Negative Gene Regulation • In a repressible operon – Binding of a specific repressor protein to the operator shuts off transcription • In an inducible operon – Binding of an inducer to an innately inactive repressor inactivates the repressor and turns on transcription • The lac operon: regulated synthesis of inducible enzymes Promoter Regulatory gene DNA Operator lacl lacZ 3 mRNA Protein No RNA made RNA polymerase 5 Active repressor (a) Lactose absent, repressor active, operon off. The lac repressor is innately active, and in the absence of lactose it switches off the operon by binding to the operator. Figure 18.22a lac operon DNA lacl lacz 3 mRNA 5 lacA RNA polymerase mRNA mRNA 55' -Galactosidase Protein Allolactose (inducer) lacY Permease Transacetylase Inactive repressor (b) Lactose present, repressor inactive, operon on. Allolactose, an isomer of lactose, derepresses the operon by inactivating the repressor. In this way, the enzymes for lactose utilization are induced. Figure 18.22b • Inducible enzymes – Usually function in catabolic pathways • Repressible enzymes – Usually function in anabolic pathways • Regulation of both the trp and lac operons – Involves the negative control of genes, because the operons are switched off by the active form of the repressor protein Positive Gene Regulation • Some operons are also subject to positive control – Via a stimulatory activator protein, such as catabolite activator protein (CAP) • In E. coli, when glucose, a preferred food source, is scarce – The lac operon is activated by the binding of a regulatory protein, catabolite activator protein (CAP) Promoter DNA lacl lacZ CAP-binding site Active CAP cAMP Inactive CAP Figure 18.23a RNA polymerase can bind and transcribe Operator Inactive lac repressor (a) Lactose present, glucose scarce (cAMP level high): abundant lac mRNA synthesized. If glucose is scarce, the high level of cAMP activates CAP, and the lac operon produces large amounts of mRNA for the lactose pathway. • When glucose levels in an E. coli cell increase – CAP detaches from the lac operon, turning it off Promoter DNA lacl lacZ CAP-binding site Operator RNA polymerase can’t bind Inactive CAP Inactive lac repressor (b) Lactose present, glucose present (cAMP level low): little lac mRNA synthesized. When glucose is present, cAMP is scarce, and CAP is unable to stimulate transcription. Figure 18.23b