* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Discussion_5_Paper_Questions
Survey
Document related concepts
Artificial gene synthesis wikipedia , lookup
Genomic library wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
DNA vaccination wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
History of genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Transcript
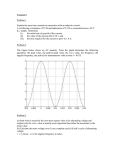
Paper Title: “An SCN9A channelopathy causes congenital inability to experience pain” Cox, 2006. http://www.nature.com/nature/journal/v444/n7121/abs/nature05413.html Purpose: The goal of this exercise is to (1) introduce students to the structure of scientific papers & (2) evidence of neurobiological research that is relevant to material they have just learned about regarding the electrophysiology of neurons and its clinical impact. Activity 1: Discuss what the purpose of each of these sections is. The structure of this paper is organized into: 1. 2. 3. 4. 5. 6. Abstract Introduction Results Discussion/Conclusion Methods Supplemental Data An overview of Nav1.7: SCN9a encodes the alpha subunit of the Nav1.7 voltage-gated Na+ channel. Together with SCN1b & SCN2b, these create a functional Nav1.7 channel. Nav1.7 is expressed in 2 tissues, nociceptive DRG neurons and sympathetic ganglion neurons. For the detection of pain, the nociceptive DRG neurons are essential. The Nav1.7 gene functions as an “amplifier” of nociception. Nociception is the detection/perception of pain, and is activated by nociceptors in nerve fibers. Nociceptors are a type of sensory receptor which responds to pain. In response to pain, nociceptors produce “generator potentials.” In this case, they are small voltage depolarizations across the neuronal membrane in DRG neurons. When there are small depolarizations from nociceptors, the Nav1.7 channel amplifies the initial generator potential, further depolarizing the neuron. If enough depolarization is reached, an AP is initiated. To clarify, the Nav1.7 does not detect the painful stimuli, it amplifies the initial detection, thus an “amplifier.” Activity 2: Questions to guide student understanding of the substance in the paper: 1. In the paper, the phenotype is an insensitivity to pain. Contemplate the “plan of attack” researchers pursued to hone in on the genetic mutations responsible for the phenotype. a. The human genome consists of billions of DNA base pairs. How can a researcher localize the mutation to a smaller region of DNA (say, to a region that is 10^6 bp, rather than 10^12)? 2. In the paper, researchers wished to examine the functional effects of the candidate mutations they identified as likely causes for the insensitivity to pain. To do this, the researchers used cell culture techniques, which involve injecting foreign DNA (plasmids) into cells growing on plates. In this paper, researchers use 2 plasmids. a. Why does the SCN9a plasmid include DsRed9 & the SCN1b/SCN2b plasmid include EGFP? Worded another way, what is the purpose of including the genes DsRed9 and EGFP on each plasmid? (reference figure 4a, 4b) b. Why, in one set of experiments, do the researchers transfect (i.e. inject DNA into cells) only the SCN1b/SCN2b plasmid into the HEK293 cells without the SCN9a? 3. Figure 4c represents the current response of a voltage clamp experiment. First, define the experimental groups. Second, explain the purpose of the experiment and the response difference between wild-type and mutants in the context of the voltage clamp experiment. 4. Figure 4d represents the current response of a voltage experiment. In Figure 4c, the voltage clamp duration is 50 ms, whereas the duration for the experiment in Figure 4d is for 500 ms. This difference is related to “voltage dependence of steady-state inactivation.” First, define the experimental group the researchers are looking at. Second, explain the chart on the right in figure 4d. A final note: Consider the knowledge base required to conduct the research presented in this paper. This includes extensive knowledge in: bioinformatics, genetics, electrophysiology, neuroanatomy, molecular biology.