* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Template for Exome Report Abstract. The abstract should include
Polycomb Group Proteins and Cancer wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Behavioural genetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene nomenclature wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Gene desert wikipedia , lookup
Essential gene wikipedia , lookup
Metagenomics wikipedia , lookup
Frameshift mutation wikipedia , lookup
X-inactivation wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Pathogenomics wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression programming wikipedia , lookup
Copy-number variation wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Public health genomics wikipedia , lookup
Genome-wide association study wikipedia , lookup
Minimal genome wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genomic imprinting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression profiling wikipedia , lookup
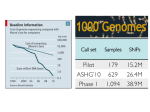
Template for Exome Report 1. Abstract. The abstract should include the list of all the genes that are discussed in the variation section 2. Clinical description. Provide full case report. Mention sex, ethnicity, parental age, biometry at birth (and gestational age), biometry at last investigation - both with centiles and/or standard deviations, psychomotor development (if possible include IQ or DQ, with the test used and at what age), phenotype at last examination, current age, and all pertinent elements of clinical history. Use the Standard Terminology proposed in Elements of Morphology or Human Phenotype Ontology (vide supra). Give pertinent family information. 3. Methods : please include Capture method Type of sequencer Variant analysis strategies (home-made pipeline, public tools…) List of databases checked (Exome Variant Server, 1000 Genomes,…) and date on which the check was done. 4. NGS quality report summary for each sample Template of the quality report table to be inserted in the manuscript. Proband Total captured regions size % of captured regions with coverage >10 Average coverage of captured region (%) Total number of SNPs Total number of INDELs 5. Add other family members (parents, sibs…) as appropriate Mb % % Variations of interest 5.1. Variations reported online All variants that fulfill one of the technical and biological criteria below must be declared in DECIPHER prior to final acceptance. The ID number of the DECIPHER data must be declared here. Declaration to other databases may be added here as well. A. Technical requirements 1. For single nucleotide variations: coverage ≥10 in proband and relatives, with raw alignment checked in 2. parents For indels: >5 variant reads and a variant/reference read ratio >0.3 B. Genetic conditions 1. 2. 3. heterozygous variants/indel present in the proband and absent in unaffected parents and controls; homozygous variants/indel and hemizygous X variants never reported in public databases genes with two rare non synonymous variants/indel with mean allelic frequency <0.03, present in the proband, but not seen together in the parents and controls The declaration is made using this DECIPHER xls file provided. All items, including those that are not mandatory for DECIPHER, must be completed, according to DECIPHER keywords. The main phenotypic traits must be added using HPO. 5.2. Subset of variations reported in the manuscript Among variants, all variants that have predicted pathogenicity (definite or probable) by at least one prediction program and occur in genes that could be hypothesized to be associated with the phenotype based on current knowledge of gene function, pathway, expression pattern and suspected inheritance mode must be presented in a tabular form (see an example below with three type of variations) and discussed. All these variants must have been checked by Sanger sequencing in the proband and the relevant relatives, and must be further discussed in the text of this section. Secondary/incidental findings in genes unrelated to the phenotype will not been reported. Template for variants of interest to be inserted in the manuscript Chromosome chr1 chr7 chr12 Position 239040006 45214519 45214604 Gene name CDC27 CES5A MST1P9 refseq NM_198317 NM_005101 NM_198576 Reference sequence A CAG T PROBAND: number of reads with reference PROBAND : alternative 12 58 0 T --- A PROBAND: number of reads with alternative 14 1 72 Other relative tested (NGS or Sanger) with the result Father : absent (exome) Mother : absent (exome) Sib II-2: absent (Sanger) Mutation type non sense Father : present (exome *+ Sanger) Mother : absent (exome + Sanger) Sib II-2: absent (Sanger) 5-UTR Father : absent (exome + Sanger) Mother : absent (exome + Sanger) Sib II-2: absent (Sanger) intronic Mutation : DNA (HGVS nomenclature _c.) Mutation : protein (HGVS nomenclature _p.) c.549A>T c.1154+48delCAG c.329+9T>A p.Ala182* Not relevant unknown Prediction < SIFT Damaging Not predicted Tolerated Prediction < PolyPhen-2 Deleterious - - Etc… Prediction < * (add row per tool) 6. Discussion. Present arguments as to why and how the candidate genes were selected. 7. 8. Facultative sections: Acknowledgements, online databases… References. Maximum 3 references per candidate gene.