* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Exploring a Protein Structure in the RCSB PDB: T
Survey
Document related concepts
Protein moonlighting wikipedia , lookup
Bottromycin wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein domain wikipedia , lookup
Proteolysis wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein folding wikipedia , lookup
Biochemistry wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Transcript
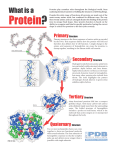
Level: Introductory Exploring a Protein Structure in the RCSB PDB: T-Cell Receptor Learning Goals: 1. Visualize the structure of a given molecule using RCSB PDB resources. 2. Explore the structure to understand its structure function relationships Exercise: Review the Molecule of the Month feature on T-cell Receptors (TCR) for background information (http://pdb101.rcsb.org/motm/63). Discuss main ideas of this feature with the students. Note that there are a few PDB entries listed throughout the feature. For example, PDB entry 1tcr, in the “Antibody Arms” section can be linked from Click on this to open the summary page for the PDB entry 1tcr (http://www.rcsb.org/pdb/explore/explore.do?structureId=1tcr). Read the provided description here and answer the following questions: 1. What is the source (organism) of the T-Cell Receptor molecule in this structure? 2. Name the authors who solved the structure of this protein? 3. Explore the 3-D structure of this protein by clicking on PV (hyperlink) next to 3D View as shown below: The default view is colored by chain (i.e. each protein (polymer) chain in the structure is colored in a different color). Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Level: Introductory Based on the 3-D model that you see here, describe the overall composition and organization of chains in the TCR structure. How many different protein chains do you see in this structure? 4. Change the viewer to JSmol using the pull-down menu. View the polymer chains shown to contain helical ribbons (in magenta), beta strand arrows (in golden yellow) and coil-like regions (white/grey). What is the predominant secondary structural element that you see here? 5. Portions of the structure are shown in ball and stick representation. What are they? What is their role in the structure? Developed as part of the RCSB Collaborative Curriculum Development Program 2015 Level: Introductory 6. Click on Custom View to see the options: Click on the box next to SS Bonds and notice yellow bond lines appear in the model. These bonds are formed by oxidation of two specific sulfur-containing amino acids. Rotate the model to visualize these bonds closely. Describe what, if any, role these bonds play in holding the TCR structure together. Developed as part of the RCSB Collaborative Curriculum Development Program 2015